
Chinese Journal OF Rice Science ›› 2024, Vol. 38 ›› Issue (4): 364-374.DOI: 10.16819/j.1001-7216.2024.231215
• Research Papers • Previous Articles Next Articles
XU Danjie1,2,3, LIN Qiaoxia1,2,3, LI Zhengkang1,2,3, ZHUANG Xiaoqian1,2,3, LING Yu1,2,3, LAI Meiling1,2,3, CHEN Xiaoting1,2,3,*( ), LU Guodong2
), LU Guodong2
Received:2023-12-15
Revised:2024-03-10
Online:2024-07-10
Published:2024-07-11
Contact:
*email: xiaotingchen@fafu.edu.cn
许丹洁1,2,3, 林巧霞1,2,3, 李正康1,2,3, 庄小倩1,2,3, 凌宇1,2,3, 赖美玲1,2,3, 陈晓婷1,2,3,*( ), 鲁国东2
), 鲁国东2
通讯作者:
*email: xiaotingchen@fafu.edu.cn
基金资助:XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance[J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374.
许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2024.231215
| 引物名称 Primer name | 序列(5’→3’) Primer sequence(5’→3’) | 用途 Purpose |
|---|---|---|
| OsOPR10-F | ATGAAGTCAAGTCCAAATCAGCTG | CDS扩增 |
| OsOPR10-R | ACGAGTTGCTAGTTCTGAATTCTGAT | CDS amplification |
| OsOPR10-OE-F | CTTCTGCAGCCCGGGGGATCCATGAAGTCAAGTCCAAATCAGCTG | 过表达载体构建 |
| OsOPR10-OE-R | CGATCGGGGAAATTCGGATCCTCAAAGATCTTCTTCAGAAATCAACTTTTGTTCACGAGTTGCTAGTTCTGAATTCTGAT | Construction of overexpression vector |
| UbiP-seq | TTTTAGCCCTGCCTTCATACGC | 过表达载体测序 |
| NosR-seq | AGACCGGCAACAGGATTCAATC | Overexpression vector sequencing |
| OsOPR10-gRT1 | CGGCCGCCTGCTTCCGTTCGTTTTAGAGCTAGAAAT | CRISPR/Cas9第一个靶点扩增 |
| OsOPR10-OsU3T1 | GAACGGAAGCAGGCGGCCGTGCCACGGATCATCTGC | Amplification of the first CRISPR/Cas9 target |
| OsOPR10-gRT2 | AGATCGGCGCGCACCGCCTGTTTTAGAGCTAGAAAT | CRISPR/Cas9第二个靶点扩增 |
| OsOPR10-U6aT2 | AGGCGGTGCGCGCCGATCTCGGCAGCCAAGCCAGCA | Amplification of the second CRISPR/Cas9 target |
| OsOPR10-T1-F | AGAGAACCGGTGCCGCTTC | CRISPR/Cas9第一个靶点验证 |
| OsOPR10-T1-R | GGTCGGAGTCGTGGCAGTC | First CRISPR/Cas9 target validation |
| Actin-QF | GAGTATGATGAGTCGGGTCCAG | Actin 定量 PCR |
| Actin-QR | ACACCAACAATCCCAAACAGAG | qRT-PCR of Actin |
| OsUG-F | TTCTGGTCCTTCCACTTTCAG | 泛素融合蛋白定量PCR |
| OsUG-R | ACGATTGATTTAACCAGTCCATGA | qRT-PCR of ubiquitin fusion protein OsUG |
| MoPot2-F | ACGACCCGTCTTTACTTATTTGG | MoPot2 定量 PCR |
| MoPot2-R | AAGTAGCGTTGGTTTTGTTGGAT | qRT-PCR of MoPot2 |
| OsOPR10-BD-F | TCAGAGGAGGACCTGCATATGATGAAGTCAAGTCCAAATCAGCTG | OsOPR10-BD载体构建 |
| OsOPR10-BD-R | TCGACGGATCCCCGGGAATTCTCAACGAGTTGCTAGTTCTGAATTC | Construction of OsOPR10-BD vector |
| OsCYP28-AD-F | GTACCAGATTACGCTCATATGATGGTGTTGCCTTCATCAAA | OsCYP28-AD载体构建 |
| OsCYP28-AD-R | ATGCCCACCCGGGTGGAATTCTTATGGCAGACTTGGAGGGG | Construction of OsCYP28-AD vector |
| OsOPR10-pGDG-F | gcttcgaattctgcagtcgacATGAAGTCAAGTCCAAATCAGCTG | OsOPR10-pGDG载体构建 |
| OsOPR10-pGDG-R | ttatctagatccggtggatccTCAACGAGTTGCTAGTTCTGAATTC | Construction of OsOPR10-pGDG vector |
| OsOPR10-PHF223-F | CGGGATCGATGAAGTCAAGTCCAAATCAG | OsOPR10-PHF223载体构建 |
| OsOPR10-PHF223-R | GGGGTACCACGAGTTGCTAGTTCTGAATT | Construction of OsOPR10-PHF223 vector |
| OsPR1a-QF | CGTCTTCATCACCTGCAACTACTC | OsPR1a 定量 PCR |
| OsPR1a-QR | CATGCATAAACACGTAGCATAGCA | qRT-PCR of OsPR1a |
| OsAOS2-QF | CAATACGTGTACTGGTCGAATGG | OsAOS2 定量 PCR |
| OsAOS2-QR | AAGGTGTCGTACCGGAGGAA | qRT-PCR of OsAOS2 |
| OsPAL1-QF | AGGAGCTCGGCTGCGTATT | OsPAL1 定量 PCR |
| OsPAL1-QR | ATGCCGAGGAACACCTTGTT | qRT-PCR of OsPAL1 |
| OsAOC-QF | CCAAGGTGCAGGAGATGTT | OsAOC 定量 PCR |
| OsAOC-QR | TACAGCTTGTTGGTGAAGGG | qRT-PCR of OsAOC |
| OsPBZ1-QF | CCCTGCCGAATACGCCTAA | OsPBZ1 定量 PCR |
| OsPBZ1-QR | CTCAAACGCCACGAGAATTTG | qRT-PCR of OsPBZ1 |
Table 1. Primers used in this study
| 引物名称 Primer name | 序列(5’→3’) Primer sequence(5’→3’) | 用途 Purpose |
|---|---|---|
| OsOPR10-F | ATGAAGTCAAGTCCAAATCAGCTG | CDS扩增 |
| OsOPR10-R | ACGAGTTGCTAGTTCTGAATTCTGAT | CDS amplification |
| OsOPR10-OE-F | CTTCTGCAGCCCGGGGGATCCATGAAGTCAAGTCCAAATCAGCTG | 过表达载体构建 |
| OsOPR10-OE-R | CGATCGGGGAAATTCGGATCCTCAAAGATCTTCTTCAGAAATCAACTTTTGTTCACGAGTTGCTAGTTCTGAATTCTGAT | Construction of overexpression vector |
| UbiP-seq | TTTTAGCCCTGCCTTCATACGC | 过表达载体测序 |
| NosR-seq | AGACCGGCAACAGGATTCAATC | Overexpression vector sequencing |
| OsOPR10-gRT1 | CGGCCGCCTGCTTCCGTTCGTTTTAGAGCTAGAAAT | CRISPR/Cas9第一个靶点扩增 |
| OsOPR10-OsU3T1 | GAACGGAAGCAGGCGGCCGTGCCACGGATCATCTGC | Amplification of the first CRISPR/Cas9 target |
| OsOPR10-gRT2 | AGATCGGCGCGCACCGCCTGTTTTAGAGCTAGAAAT | CRISPR/Cas9第二个靶点扩增 |
| OsOPR10-U6aT2 | AGGCGGTGCGCGCCGATCTCGGCAGCCAAGCCAGCA | Amplification of the second CRISPR/Cas9 target |
| OsOPR10-T1-F | AGAGAACCGGTGCCGCTTC | CRISPR/Cas9第一个靶点验证 |
| OsOPR10-T1-R | GGTCGGAGTCGTGGCAGTC | First CRISPR/Cas9 target validation |
| Actin-QF | GAGTATGATGAGTCGGGTCCAG | Actin 定量 PCR |
| Actin-QR | ACACCAACAATCCCAAACAGAG | qRT-PCR of Actin |
| OsUG-F | TTCTGGTCCTTCCACTTTCAG | 泛素融合蛋白定量PCR |
| OsUG-R | ACGATTGATTTAACCAGTCCATGA | qRT-PCR of ubiquitin fusion protein OsUG |
| MoPot2-F | ACGACCCGTCTTTACTTATTTGG | MoPot2 定量 PCR |
| MoPot2-R | AAGTAGCGTTGGTTTTGTTGGAT | qRT-PCR of MoPot2 |
| OsOPR10-BD-F | TCAGAGGAGGACCTGCATATGATGAAGTCAAGTCCAAATCAGCTG | OsOPR10-BD载体构建 |
| OsOPR10-BD-R | TCGACGGATCCCCGGGAATTCTCAACGAGTTGCTAGTTCTGAATTC | Construction of OsOPR10-BD vector |
| OsCYP28-AD-F | GTACCAGATTACGCTCATATGATGGTGTTGCCTTCATCAAA | OsCYP28-AD载体构建 |
| OsCYP28-AD-R | ATGCCCACCCGGGTGGAATTCTTATGGCAGACTTGGAGGGG | Construction of OsCYP28-AD vector |
| OsOPR10-pGDG-F | gcttcgaattctgcagtcgacATGAAGTCAAGTCCAAATCAGCTG | OsOPR10-pGDG载体构建 |
| OsOPR10-pGDG-R | ttatctagatccggtggatccTCAACGAGTTGCTAGTTCTGAATTC | Construction of OsOPR10-pGDG vector |
| OsOPR10-PHF223-F | CGGGATCGATGAAGTCAAGTCCAAATCAG | OsOPR10-PHF223载体构建 |
| OsOPR10-PHF223-R | GGGGTACCACGAGTTGCTAGTTCTGAATT | Construction of OsOPR10-PHF223 vector |
| OsPR1a-QF | CGTCTTCATCACCTGCAACTACTC | OsPR1a 定量 PCR |
| OsPR1a-QR | CATGCATAAACACGTAGCATAGCA | qRT-PCR of OsPR1a |
| OsAOS2-QF | CAATACGTGTACTGGTCGAATGG | OsAOS2 定量 PCR |
| OsAOS2-QR | AAGGTGTCGTACCGGAGGAA | qRT-PCR of OsAOS2 |
| OsPAL1-QF | AGGAGCTCGGCTGCGTATT | OsPAL1 定量 PCR |
| OsPAL1-QR | ATGCCGAGGAACACCTTGTT | qRT-PCR of OsPAL1 |
| OsAOC-QF | CCAAGGTGCAGGAGATGTT | OsAOC 定量 PCR |
| OsAOC-QR | TACAGCTTGTTGGTGAAGGG | qRT-PCR of OsAOC |
| OsPBZ1-QF | CCCTGCCGAATACGCCTAA | OsPBZ1 定量 PCR |
| OsPBZ1-QR | CTCAAACGCCACGAGAATTTG | qRT-PCR of OsPBZ1 |

Fig. 1. Expression patterns of OsOPR10 after MeJA (A) and SA (B) application Data in the figure are Mean±SD, and *, ** and ***represent significant difference at the 0.05, 0.01 and 0.001 levels (t-test), respectively. The same below.

Fig. 3. Knockout target design and mutation type identification of OsOPR10 A, Two gRNA targets of OsOPR10 gene; B, Detection of mutantion sites in OsOPR10 knockout transgenic line.

Fig. 5. Response of OsOPR10 transgenic rice to M. oryzae A, Lesions after inoculation with M. oryzae; B, Lesion area after inoculation with M. oryzae; C, Relative fungal biomass after inoculation with M. oryzae. Data in the figure are Mean±SD, and *** represent significant difference at 0.001 level(t-test), respectively, n=4.

Fig. 6. Response of OsOPR10 transgenic rice to Xoo. A, Lesions size after inoculation by Xoo by leaf clipping method; B, Length of lesions after inoculation with Xoo. Data in the figure are Mean±SD, and * represents significant difference at the 0.05 levels (t-test), n=5.

Fig. 7. Response of OsOPR10 transgenic rice to chitin and flg22 induction. A, Chitin-induced ROS burst in OsOPR10-OE transgenic rice and Nipponbare; B, Chitin-induced ROS burst in OsOPR10-KO transgenic rice and Nipponbare; C, flg22-induced ROS burst in OsOPR10-OE transgenic rice and Nipponbare; D, flg22-induced ROS burst in OsOPR10-KO transgenic rice and Nipponbare.
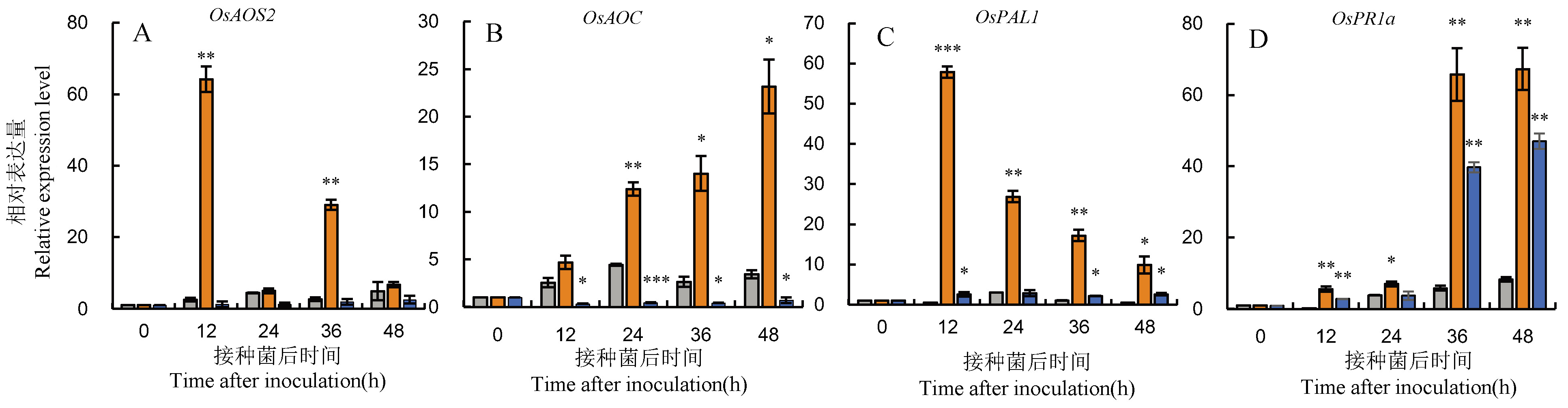
Fig. 8. Expression analysis of genes involved in JA and SA pathways in OsOPR10 transgenic rice after infection by M. oryzae Data in the figure are Mean±SD, *, ** and *** represent significant difference at the 0.05, 0.01 and 0.001 levels (t-test), respectively.

Fig. 9. Subcellular localization of OsOPR10-PHF223 and OsOPR10-pGDG fusion protein A, Subcellular localization of OsOPR10-PHF223 fusion protein and PHF223 vector in rice protoplasts, the scale bar in the figure represents 10 μm; B, Subcellular localization of OsOPR10-pGDG fusion protein and pGDG vector in tobacco leaves, the scale bar in the figure represents 25 μm.

Fig. 11. Luciferase complementation assay verifies the interaction between OsOPR10 and OsCYP28 a: OsOPR10-Nluc+OsCYP28-Cluc; b: OsOPR10-Nluc+Cluc; c: OsCYP28-Cluc+Nluc; d: Nluc+Cluc.
| [1] | Ainsworth E A. Rice production in a changing climate: A meta-analysis of responses to elevated carbon dioxide and elevated ozone concentration[J]. Global Change Biology, 2010, 14(7): 1642-1650. |
| [2] | Dean R, Kan J, Pretorius Z A, Hammond-Kosack K E, Pietro A D, Spanu P D, Ruddd J J, Pickman M. The Top 10 fungal pathogens in molecular plant pathology[J]. Molecular Plant Pathology, 2012, 13(4): 414-430. |
| [3] | Jiang N, Yan J, Liang Y, Shi Y L, He Z Z, Wu Y T, Zeng Q, Liu X L, Peng J H. Resistance genes and their interactions with bacterial blight pathogens in rice[J]. Rice, 2020, 13(1): 1-12. |
| [4] | Liu W D, Wang G L. Plant innate immunity in rice: A defense against pathogen infection[J]. National Science Review, 2016, 3: 295-308. |
| [5] | Yuan M H, Jiang Z Y, Bi G Z, Nomura K Y, Liu M H, Wang Y P, Cai B Y, Zhou J M, He S Y, Xin X F. Pattern- recognition receptors are required for NLR-mediated plant immunity[J]. Nature, 2021, 592(7852): 105-109. |
| [6] | 覃磊, 彭志红, 夏石头. 植物NLR免疫受体的识别、免疫激活与信号调控[J]. 植物学报, 2022, 57(1): 12-23. |
| Qin L, Peng Z H, Xia S T. Recognition, immune activation and signal regulation of plant NLR immune receptor[J]. Chinese Bulletin of Botany, 2022, 57(1): 12-23. (in Chinese with English abstract) | |
| [7] | Ramírez-Zavaleta C Y, García-Barrera L J, Rodríguez- Verástegui L L, Arrieta-Flores D. An overview of PRR-and NLR-mediated immunities: Conserved signaling components across the plant kingdom that communicate both pathways[J]. International Journal of Molecular Sciences, 2022, 23(21): 12974. |
| [8] | Qiu J, Xie J, Chen Y, Shen Z, Shi H, Naqvi N I, Qian Q, Liang Y, Kou Y J. Warm temperature compromises JA-regulated basal resistance to enhance Magnaporthe oryzae infection in rice[J]. Molecular Plant, 2022, 15(4): 723-739. |
| [9] | Wasternack C, Hause B. Jasmonates and octadecanoids: Signals in plant stress responses and development[J]. Progress in Nucleic Acid Research and Molecular Biology, 2002, 72: 165-221. |
| [10] | Liechti R. The jasmonate pathway[J]. Science, 2002, 296(5573): 1649-1650. |
| [11] | Al-Momany B, Abu-Romman S. Homologs of old yellow enzyme in plants[J]. Australian Journal of Crop Science. 2016, 10(4): 584-590. |
| [12] | Agrawal G K, Jwa N S, Shibato J, Han O, Iwahashi H, Rakwal R. Diverse environmental cues transiently regulate OsOPR1 of the octadecanoid pathway revealing its importance in rice defense/stress and development[J]. Biochemical and Biophysical Research Communications, 2003, 310(4): 1073-1082. |
| [13] | Guo H M, Li H C, Zhou S R, Xue H W, Miao X X. Cis-12-oxo-phytodienoic acid stimulates rice defense response to a piercing-sucking insect[J]. Molecular Plant, 2014, 7(11): 1683-1692. |
| [14] | Tani T, Sobajima H, Okada K, Chujo T, Arimura S, Tsutsumi N, Nishimura M, Seto H, Nojiri H, Yamane H. Identification of OsOPR7 encoding 12-oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice: An international journal of plant biology[J]. Planta, 2008, 227(3): 517-526. |
| [15] | Cong L, Ran F A, Cox D, Lin S L, Barretto R, Habib N, Hsu P D, Wu X, Jiang W Y, Marraffini L A, Zhang F. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823. |
| [16] | Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V. Editing plant genomes with CRISPR/ Cas9[J]. Current Opinion in Biotechnology, 2015, 32: 76-84. |
| [17] | 曾栋昌, 马兴亮, 谢先荣, 祝钦泷, 刘耀光. 植物CRISPR/Cas9多基因编辑载体构建和突变分析的操作方法[J]. 中国科学: 生命科学, 2018, 48(7): 12. |
| Zeng D C, Ma X L, Xie X R, Zhu Q L, Liu Y G. Operational methods for the construction of plant CRISPR/Cas9 multi-gene editing vectors and mutation analysis[J]. Science China: Life Sciences, 2018, 48(7): 12. (in Chinese with English abstract) | |
| [18] | Clarke J D. Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation[J]. Cold Spring Harbor Protocol, 2009(3): pdb.prot5177. |
| [19] | Shi X T, Long Y, He F, Zhang C Y, Wang R Y, Zhang T, Wu W, Hao Z Y, Wang Y, Wang G L, Ning Y S. The fungal pathogen Magnaporthe oryzae suppresses innate immunity by modulating a host potassium channel[J]. PLoS Pathogens, 2018, 14(1): e1006878. |
| [20] | Hao Z Y, Tian J F, Fang H, Fang L, Xu X, He F, Li S Y, Xie W Y, Du Q, You X M, Wang D B, Chen Q H, Wang R Y, Zuo S M. A VQ-motif-containing protein fine-tunes rice immunity and growth by a hierarchical regulatory mechanism[J]. Cell Reports, 2022, 40(7): 111235. |
| [21] | Park C H, Chen S, Shirsekar G, Zhou B, Khang C H, Songkumarn P, Afzal A J, Ning Y S, Wang R Y, Bellizzi M, Valent B, Wang G L. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice[J]. Plant Cell, 2012, 24(11): 4748-4762. |
| [22] | Yoo S D, Cho Y H, Sheen J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis[J]. Nature Protocols, 2007, 2(7): 1565-1572. |
| [23] | Li R L, Wang J L, Xu L, Sun M H, Yi K K, Zhao H Y. Functional analysis of phosphate transporter OsPHT4 family members[J]. Rice Science, 2020, 27(6): 493-503. |
| [24] | Gu Y H, Li G N, Wang P, Guo Y, Li J R. A simple and precise method (Y2H-in-frame-seq) improves yeast two-hybrid screening with cDNA libraries[J]. Journal of Genetics Genomics, 2022, 49(6): 595-598. |
| [25] | Mou Y F, Liu Y Y, Tian S J, Guo Q P, Wang C S, Wen S S. Genome-Wide identification and characterization of the OPR gene family in wheat[J]. International Journal of Molecular Sciences, 2019, 20(8): 1914. |
| [26] | Zhang J L, Simmons C, Yalpani N, Crane V, Wilkinson H, Kolomiets M. Genomic analysis of the 12-oxo- phytodienoic acid reductase gene family of Zea mays[J]. Plant Molecular Biology, 2005, 59: 323-334. |
| [27] | Piñera-Chavez F J, Berry P M, Foulkes M J, Molero G, Reynolds M P. Avoiding lodging in irrigated spring wheat. II. Genetic variation of stem and root structural properties[J]. Field Crops Research, 2016, 196: 64-74. |
| [28] | Scalschi L, Sanmartín M, Camañes G, Troncho P, Sánchez-Serrano J J, García-Agustín P, Vicedo B. Silencing of OPR3 in tomato reveals the role of OPDA in callose deposition during the activation of defense responses against Botrytis cinerea[J]. Plant Journal, 2015, 81(2): 304-315. |
| [29] | Li C, Xu M X, Cai X, Han Z G, Si J P, Chen D H. Jasmonate signaling pathway modulates plant defense, growth, and their trade-offs[J]. International Journal of Molecular Sciences, 2022, 23(7): 3945. |
| [30] | 文锴, 王远, 胡蝶, 袁敬平, 侯喜林, 李英. 不结球白菜BcOPR3基因的克隆与功能分析[J]. 南京农业大学学报, 2017, 40(5): 8. |
| Wen K, Wang Y, Hu D, Yuan J P, Hou X L, Li Y. Cloning and expression analysis of BcOPR3 gene in non-heading Chinese cabbage[J]. Journal of Nanjing Agricultural University, 2017, 40(5): 8. (in Chinese with English abstract) | |
| [31] | 王艳微, 王敏, 王江, 张庆祝, 解莉楠. 大豆OPR基因家族全基因组鉴定与表达分析[J]. 大豆科学, 2022, 41(2): 129-139. |
| Wang Y W, Wang M, Wang J, Zhang Q Z, Xie L N. Genome-wide identification and expression analysis of soybean OPR gene family[J]. Soybean Science, 2022, 41(2): 129-139. (in Chinese with English abstract) | |
| [32] | Dong W, Wang M C, Xu F, Quan T Y, Peng K Q, Xiao L T, Xia G M. Wheat oxophytodienoate reductase gene TaOPR1 confers salinity tolerance via enhancement of abscisic acid signaling[J]. Plant Physiology, 2013, 161(3): 1217-1228. |
| [33] | Wang Y K, Yuan G L, Yuan S H, Duan W J, Wang P, Bai J F, Zhang F T, Gao S Q, Zhang L P, Zhao C P. TaOPR2 encodes a 12-oxo-phytodienoic acid reductase involved in the biosynthesis of jasmonic acid in wheat (Triticum aestivum L.)[J]. Biochemical and Biophysical Research Communications, 2016, 470(1): 233-238. |
| [34] | 游双红, 谭平, 史文景, 武峥, 陈元平, 伊洪伟, 周广文. 葡萄OPR家族全基因组鉴定及重金属胁迫下的表达分析[J]. 基因组学与应用生物学, 2021, 40(5): 10. |
| You S H, Tan P, Shi W J, Wu Z, Chen Y P, Yi H W, Zhou G W. Genome-wide identification of OPR family genes and the expression of these genes in response to heavy metal stress[J]. Genomics and Applied Biology, 2021, 40(5): 10. (in Chinese with English abstract) | |
| [35] | Li M P, Kim C H. Chloroplast ROS and stress signaling[J]. Plant Communication, 2021, 3(1): 100264. |
| [36] | Gao H X, Zhu L, Liu T Q, Leng X Y, Zhu Z X, Xie W, Lü H T, Jin Z X, Wu P, Zhang Z C. Identification of a novel OsCYP2 allele that was involved in rice response to low temperature stress[J]. Phyton-International Journal of Experimental Botany, 2023, 92(6): 1743-1763. |
| [37] | Zhu W N, Xu L Q, Yu X X, Zhong Y. The immunophilin CYCLOPHILIN28 affects PSII-LHCII supercomplex assembly and accumulation in Arabidopsis thaliana[J]. Journal of Integrative Plant Biology, 2022, 64(4): 915-929. |
| [1] |
WANG Yichen, ZHU Benshun, ZHOU Lei, ZHU Jun, YANG Zhongnan.
Sterility Mechanism of Photoperiod/Thermo-sensitive Genic Male Sterile Lines and Development and Prospects of Two-line Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(5): 463-474. |
| [2] |
XU Yongqiang XU Jun, FENG Baohua, XIAO Jingjing, WANG Danying, ZENG Yuxiang, FU Guanfu.
Research Progress of Pollen Tube Growth in Pistil of Rice and Its Response to Abiotic stress [J]. Chinese Journal OF Rice Science, 2024, 38(5): 495-506. |
| [3] |
HE Yong, LIU Yaowei, XIONG Xiang, ZHU Danchen, WANG Aiqun, MA Lana, WANG Tingbao, ZHANG Jian, LI Jianxiong, TIAN Zhihong.
Creation of Rice Grain Size Mutants by Editing OsOFP30 via CRISPR/Cas9 System [J]. Chinese Journal OF Rice Science, 2024, 38(5): 507-515. |
| [4] |
LÜ Yang, LIU Congcong, YANG Longbo, CAO Xinglan, WANG Yueying, TONG Yi, Mohamed Hazman, QIAN Qian, SHANG Lianguang, GUO Longbiao.
Identification of Candidate Genes for Rice Nitrogen Use Efficiency by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(5): 516-524. |
| [5] |
YANG Hao, HUANG Yanyan, WANG Jian, YI Chunlin, SHI Jun, TAN Chutian, REN Wenrui, WANG Wenming.
Development and Application of Specific Molecular Markers for Eight Rice Blast Resistance Genes in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(5): 525-534. |
| [6] |
JIANG Peng, ZHANG Lin, ZHOU Xingbing, GUO Xiaoyi, ZHU Yongchuan, LIU Mao, GUO Chanchun, XIONG Hong, XU Fuxian.
Yield Formation Characteristics of Ratooning Hybrid Rice Under Simplified Cultivation Practices in Winter Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(5): 544-554. |
| [7] |
YANG Mingyu, CHEN Zhicheng, PAN Meiqing, ZHANG Bianhong, PAN Ruixin, YOU Lindong, CHEN Xiaoyan, TANG Lina, HUANG Jinwen.
Effects of Nitrogen Reduction Combined with Biochar Application on Stem and Sheath Assimilate Translocation and Yield Formation in Rice Under Tobacco-rice Rotation [J]. Chinese Journal OF Rice Science, 2024, 38(5): 555-566. |
| [8] |
XIONG Jiahuan, ZHANG Yikai, XIANG Jing, CHEN Huizhe, XU Yicheng, WANG Yaliang, WANG Zhigang, YAO Jian, ZHANG Yuping .
Effect of Biochar-based Fertilizer Application on Rice Yield and Nitrogen Utilization in Film- mulched PaddyFields [J]. Chinese Journal OF Rice Science, 2024, 38(5): 567-576. |
| [9] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [10] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [11] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [12] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [13] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [14] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [15] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||