
Chinese Journal OF Rice Science ›› 2024, Vol. 38 ›› Issue (3): 266-276.DOI: 10.16819/j.1001-7216.2024.230904
• Research Papers • Previous Articles Next Articles
ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing*( ), HE Jiwai*(
), HE Jiwai*( )
)
Received:2023-09-12
Revised:2023-12-29
Online:2024-05-10
Published:2024-05-13
Contact:
*email: hunanhongli@aliyun.com;
hejiwai@hunau.edu.cn
朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清*( ), 贺记外*(
), 贺记外*( )
)
通讯作者:
*email: hunanhongli@aliyun.com;
hejiwai@hunau.edu.cn
基金资助:ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis[J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276.
朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2024.230904
| 年份 Year | QTL | 染色体 Chr | 关联位点 SNP | 位置 Position | P值 P value | 贡献率 R2(%) |
|---|---|---|---|---|---|---|
| 2020 | qTA2 | 2 | 78 251 484 | 34 980 561 | 2.82×10−5 | 13.28 |
| qTA9 | 9 | 290 983 508 | 20 510 958 | 2.58×10−5 | 13.66 | |
| qTA12 | 12 | 365 807 909 | 20 094 246 | 3.08×10−6 | 16.22 | |
| 2022 | qTA5 | 5 | 165 535 507 | 14 410 821 | 4.36×10−5 | 6.23 |
| qTA6.1 | 6 | 208 648 428 | 27 565 308 | 1.95×10−6 | 8.29 | |
| qTA6.2 | 6 | 209 509 442 | 28 426 322 | 7.28×10−6 | 7.54 | |
| qTA9 | 9 | 291 032 810 | 20 560 260 | 7.45×10−8 | 10.41 |
Table 1. List of QTL for tiller angle by GWAS
| 年份 Year | QTL | 染色体 Chr | 关联位点 SNP | 位置 Position | P值 P value | 贡献率 R2(%) |
|---|---|---|---|---|---|---|
| 2020 | qTA2 | 2 | 78 251 484 | 34 980 561 | 2.82×10−5 | 13.28 |
| qTA9 | 9 | 290 983 508 | 20 510 958 | 2.58×10−5 | 13.66 | |
| qTA12 | 12 | 365 807 909 | 20 094 246 | 3.08×10−6 | 16.22 | |
| 2022 | qTA5 | 5 | 165 535 507 | 14 410 821 | 4.36×10−5 | 6.23 |
| qTA6.1 | 6 | 208 648 428 | 27 565 308 | 1.95×10−6 | 8.29 | |
| qTA6.2 | 6 | 209 509 442 | 28 426 322 | 7.28×10−6 | 7.54 | |
| qTA9 | 9 | 291 032 810 | 20 560 260 | 7.45×10−8 | 10.41 |
| QTL位点 QTL | 候选基因 Candidate gene | 功能注释 Description |
|---|---|---|
| qTA2 | Os02g0817600 | 生长素/吲哚-3-乙酸蛋白 |
| Auxin/indole-3-acetic acid protein | ||
| Os02g0817900 | 细胞色素P450家族蛋白 | |
| Cytochrome P450 family protein | ||
| qTA6.1 | Os06g0663800 | 叶绿体前体 |
| Chloroplast precursor | ||
| Os06g0665400 | F-box结构域蛋白 | |
| F-box domain containing protein | ||
| Os06g0666500 | 锌指结构域蛋白 | |
| Zinc finger domain containing protein | ||
| Os06g0679400 | Myb转录因子 | |
| Myb-transcription factor | ||
| qTA6.2 | Os06g0680700 | 细胞色素P450家族蛋白 |
| Cytochrome P450 family protein. | ||
| Os06g0682800 | 锌指结构域蛋白 | |
| Zinc finger domain containing protein | ||
| Os06g0683000 | 锌指结构域蛋白 | |
| Zinc finger domain containing protein | ||
| Os12g0516900 | F-box结构域蛋白 | |
| F-box domain containing protein | ||
| qTA12 | Os12g0517000 | F-box结构域蛋白 |
| F-box domain containing protein | ||
| Os12g0517100 | F-box结构域蛋白 | |
| F-box domain containing protein |
Table 2. Candidate genes of rice tiller angle
| QTL位点 QTL | 候选基因 Candidate gene | 功能注释 Description |
|---|---|---|
| qTA2 | Os02g0817600 | 生长素/吲哚-3-乙酸蛋白 |
| Auxin/indole-3-acetic acid protein | ||
| Os02g0817900 | 细胞色素P450家族蛋白 | |
| Cytochrome P450 family protein | ||
| qTA6.1 | Os06g0663800 | 叶绿体前体 |
| Chloroplast precursor | ||
| Os06g0665400 | F-box结构域蛋白 | |
| F-box domain containing protein | ||
| Os06g0666500 | 锌指结构域蛋白 | |
| Zinc finger domain containing protein | ||
| Os06g0679400 | Myb转录因子 | |
| Myb-transcription factor | ||
| qTA6.2 | Os06g0680700 | 细胞色素P450家族蛋白 |
| Cytochrome P450 family protein. | ||
| Os06g0682800 | 锌指结构域蛋白 | |
| Zinc finger domain containing protein | ||
| Os06g0683000 | 锌指结构域蛋白 | |
| Zinc finger domain containing protein | ||
| Os12g0516900 | F-box结构域蛋白 | |
| F-box domain containing protein | ||
| qTA12 | Os12g0517000 | F-box结构域蛋白 |
| F-box domain containing protein | ||
| Os12g0517100 | F-box结构域蛋白 | |
| F-box domain containing protein |
| 基因 Gene | 单倍型1/种质数 Hap1/Accession number | 单倍型2/种质数 Hap2/Accession number | 单倍型3/种质数 Hap3/Accession number | |
|---|---|---|---|---|
| Os02g0817900 | AACGGACAAT/51 | AACGGGCAAT/33 | TGATGACGGCGC/81 | |
| Os06g0682800 | GGGATG/77 | GATACCA/70 | ||
| Os06g0663800 | ATGGCCCGC/46 | ATTGCCCGC/41 | GCGAATTAT/74 | |
| Os02g0817600 | CGATCC/56 | CGCGTT/25 | GACGTC/50 | |
| Os06g0680700 | AATTCAATGAAGCTGAAG/78 | GGGGGGGCAGGCGCTGGT/79 | ||
Table 3. Candidate gene haplotype group and composition of each haplotype SNP
| 基因 Gene | 单倍型1/种质数 Hap1/Accession number | 单倍型2/种质数 Hap2/Accession number | 单倍型3/种质数 Hap3/Accession number | |
|---|---|---|---|---|
| Os02g0817900 | AACGGACAAT/51 | AACGGGCAAT/33 | TGATGACGGCGC/81 | |
| Os06g0682800 | GGGATG/77 | GATACCA/70 | ||
| Os06g0663800 | ATGGCCCGC/46 | ATTGCCCGC/41 | GCGAATTAT/74 | |
| Os02g0817600 | CGATCC/56 | CGCGTT/25 | GACGTC/50 | |
| Os06g0680700 | AATTCAATGAAGCTGAAG/78 | GGGGGGGCAGGCGCTGGT/79 | ||
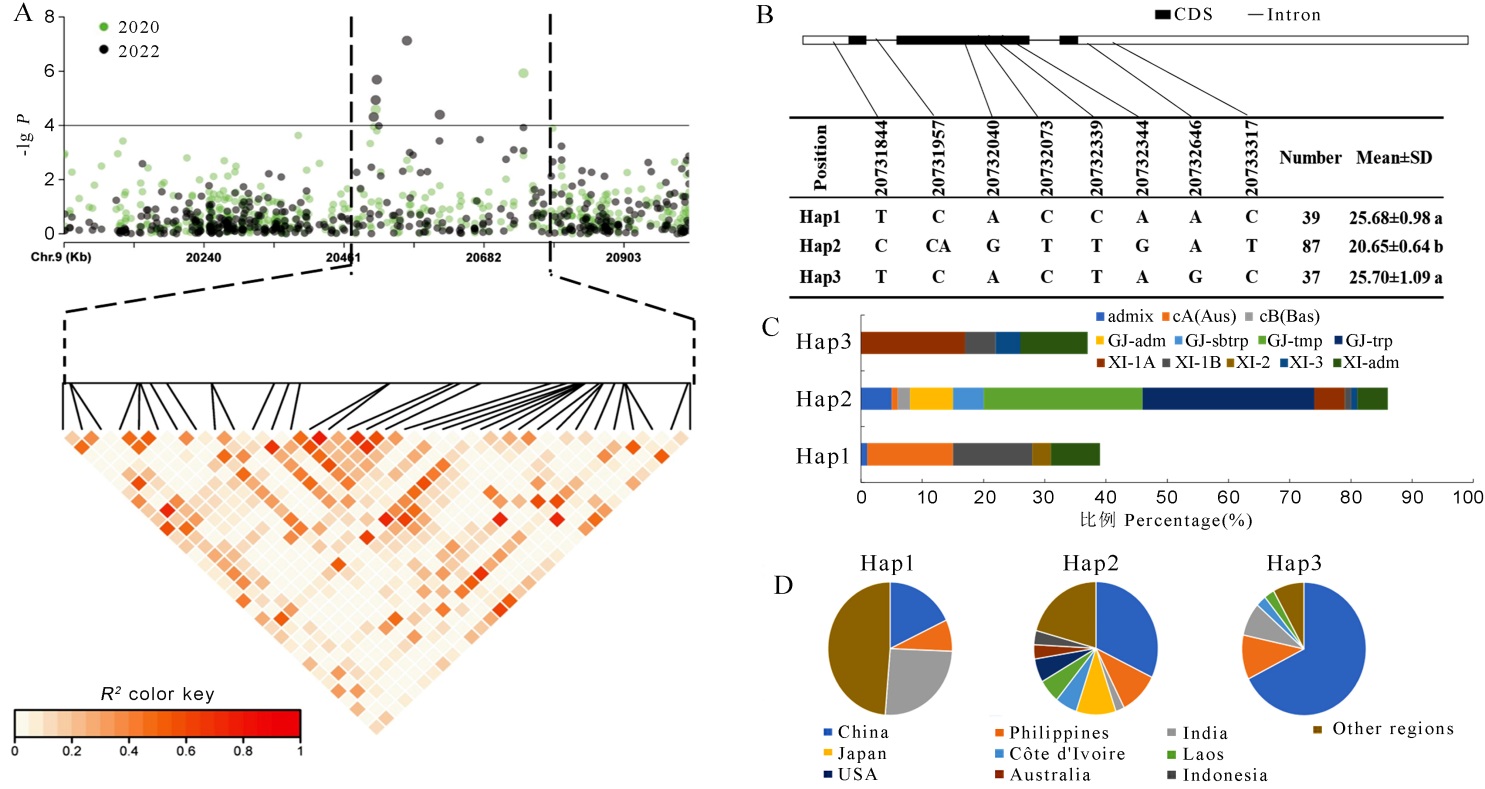
Fig. 4. TAC1 haplotype analysis A, Linkage disequilibrium plot for SNPs with -lg P> 4 in qTA9; B, TAC1 gene structure and haplotype schematic. Different lowercase letters indicate significant differences (P<0.05); C: The subpopulation composition of TAC1 haplotypes; D, Geographical distribution of different haplotypes of TAC1.
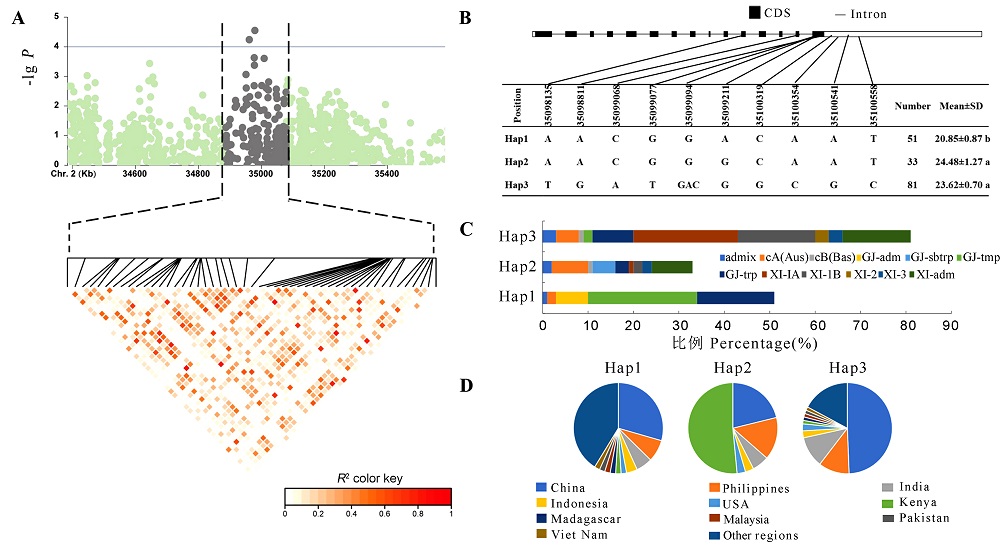
Fig. 5. Os02g0817900 haplotype analysis A, Linkage disequilibrium plot for SNPs with -lg P> 4 in qTA2; B, Os02g0817900 structure and haplotype schematic. Different lowercase letters indicate significant differences(P<0.05); C, Subpopulation composition of Os02g0817900 haplotypes; D, Geographical distribution of haplotypes of Os02g0817900.
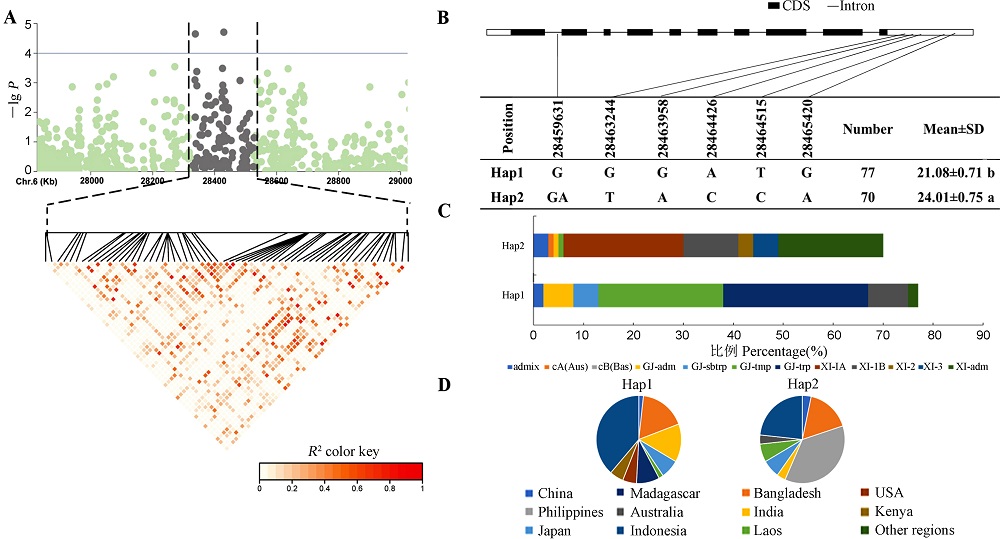
Fig. 6. Os06g0682800 haplotype analysis A, Linkage disequilibrium plot for SNPs with -lg P> 4 in qTA6; B, Os06g0682800 structure and haplotype Schematic. Different lowercase letters indicate significant differences(P<0.05); C, Subpopulation composition of Os06g0682800 haplotypes; D, Geographical distribution of haplotypes of Os06g0682800.
| [1] | 朱亮, 薛蓬勃, 李国强, 黄婧, 金健. 全基因组结合位点分析揭示LAZY1控制水稻分蘖角度的下游调控网络[J]. 基因组学与应用生物学, 2022, 41(7): 1539-1549. |
| Zhu L, Xue P B, Li G Q, Huang J, Jin J. Genome-wide binding site analysis of LAZY1 reveals its downstream regulating network in controlling rice tiller angle[J]. Genomics and Applied Biology, 2022, 41(7): 1539-1549. (in Chinese with English abstract) | |
| [2] | 黄卫衡, 吕启明, 辛业芸. 水稻分蘖角度的研究进展[J]. 杂交水稻, 2019, 34(3): 1-7. |
| Huang W H, Lü Q M, Xin Y Y. Research progress on rice tiller angle[J]. Hybrid Rice, 2019, 34(3): 1-7. (in Chinese with English abstract) | |
| [3] | 钱前, 何平, 滕胜, 曾大力, 朱立煌. 水稻分蘖角度的QTLs分析[J]. 遗传学报, 2001(1): 29-32. |
| Qian Q, He P, Teng S, Zeng D L, Zhu L H. QTL analysis on rice tiller angle[J]. Journal of Genetics and Genomics, 2001(1): 29-32. (in Chinese with English abstract) | |
| [4] | 余传元, 刘裕强, 江玲, 王春明, 翟虎渠, 万建民. 水稻分蘖角度的QTL定位和主效基因的遗传分析[J]. 遗传学报, 2005(9): 948-954. |
| Yu C Y, Liu Y Q, Jiang L, Wang C M, Zhai H Q, Wan J M. QTLs mapping and genetic analysis of tiller angle in rice[J]. Journal of Genetics and Genomics, 2005(9): 948-954. (in Chinese with English abstract) | |
| [5] | Yu B S, Lin Z W, Li H X, Li X J, Li J Y, Wang Y H, Zhang X, Zhu Z F, Zhai W X, Wang X K, Xie D X, Sun C Q. TAC1, a major quantitative trait locus controlling tiller angle in rice[J]. Plant Journal, 2007, 52(5): 891. |
| [6] | Dong H J, Zhao H, Xie W B, Han Z M, Li G W, Yao W, Bai X F, Hu Y, Guo Z L, Lu K, Yang L, Xing Y Z. A novel tiller angle gene, TAC3, together with TAC1 and D2 largely determine the natural variation of tiller angle in rice cultivars[J]. PLoS Genet, 2016, 12(11): e1006412. |
| [7] | He J W, Shao G N, Wei X J, Huang F L, Sheng Z H, Tang S Q, Hu P S. Fine mapping and candidate gene analysis of qTAC8, a major quantitative trait locus controlling tiller angle in rice (Oryza sativa L.)[J]. PLoS One, 2017, 12(5): e0178177. |
| [8] | Tan L B, Li X R, Liu F X, Sun X Y, Li C G, Zhu Z F, Fu Y C, Cai H W, Wang X K, Xie D X, Sun C Q. Control of a key transition from prostrate to erect growth in rice domestication[J]. Nature Genetics, 2008, 40(11): 1360. |
| [9] | Jin J, Huang W, Gao J P, Yang J, Shi M, Zhu M Z, Luo D, Lin H X. Genetic control of rice plant architecture under domestication[J]. Nature Genetics, 2008, 40(11): 1365-1369. |
| [10] | Zhang W F, Tan L B, Sun H Y, Zhao X H, Liu F X, Cai H W, Fu Y C, Sun X Y, Gu P, Zhu Z F, Sun C Q. Natural variations at TIG1 encoding a TCP transcription factor contribute to plant architecture domestication in rice[J]. Molecular Plant, 2019, 12(8): 1075-1089. |
| [11] | Okamura M, Hirose T, Hashida Y, Yamagishi T, Ohsugi R, Aoki N. Starch reduction in rice stems due to a lack of OsAGPL1 or OsAPL3 decreases grain yield under low irradiance during ripening and modifies plant architecture[J]. Functional Plant Biology, 2013, 40(11): 1137-1146. |
| [12] | Huang L Z, Wang W G, Zhang N, Cai Y Y, Liang Y, Meng X B, Yuan Y D, Li J Y, Wu D X, Wang Y H. LAZY2 controls rice tiller angle through regulating starch biosynthesis in gravity-sensing cells[J]. New Phytologist, 2021, 231(3): 1073-1087. |
| [13] | Sakuraba Y, Piao W, Lim J H, Han S H, Kim Y S, An G, Paek N C. Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle[J]. Plant Cell Physiology, 2015, 56(12): 2325-2339. |
| [14] | Wu X R, Tang D, Li M, Wang K J, Cheng Z K. Loose Plant Architecture1, an indeterminate domain protein involved in shoot gravitropism, regulates plant architecture in rice[J]. Plant Physiology, 2013, 161: 317. |
| [15] | Zhang N, Yu H, Yu H, Cai Y Y, Huang L Z, Xu C, Xiong G S, Meng X B, Wang J Y, Chen H F, Liu G F, Jing Y H, Yuan Y D, Liang Y, Li S J, Smith S M, Li J Y, Wang Y H. A core regulatory pathway controlling rice tiller angle mediated by the LAZY1-dependent asymmetric distribution of auxin[J]. The Plant Cell, 2018, 30(7): 1461-1475. |
| [16] | Li P J, Wang Y H, Qian Q, Fu Z M, Wang M, Zeng D L, Li B H, Wang X J, Li G Y. LAZY1 controls rice shoot gravitropism through regulating polar auxin transport[J]. Cell Research, 2007, 17(5): 402-410. |
| [17] | Yoshihara T, Iino M. Identification of the gravitropism-related rice gene LAZY1 and elucidation of LAZY1-dependent and -independent gravity signaling pathways[J]. Plant Cell Physiology, 2007, 48(5): 678. |
| [18] | Li Z, Liang Y, Yuan Y D, Wang L, Meng X B, Xiong G S, Zhou J, Cai Y Y, Han N P, Hua L K, Liu G F, Li J Y, Wang Y H. OsBRXL4 regulates shoot gravitropism and rice tiller angle through affecting LAZY1 nuclear localization[J]. Molecular Plant, 2019, 12(8): 1143-1156. |
| [19] | Hu Y, Li S L, Fan X W, Song S, Zhou X, Weng X Y, Xiao J H, Li X H, Xiong L Z, You A Q, Xing Y Z. OsHOX1 and OsHOX28 redundantly shape rice tiller angle by reducing HSFA2D expression and auxin content[J]. Plant Physiology, 2020, 184(3): 1424-1437. |
| [20] | Li Y, Li J L, Chen Z H, Wei Y, Qi Y H, Wu C Y. OsmiR167a-targeted auxin response factors modulate tiller angle via fine-tuning auxin distribution in rice[J]. Plant Biotechnology Journal, 2020, 18(10): 2015-2026. |
| [21] | Chen Y N, Fan X R, Song W J, Zhang Y L, Xu G H. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1[J]. Plant Biotechnology Journal, 2012, 10(2): 139-149. |
| [22] | Zhao L, Tan L B, Zhu Z F, Xiao L T, Xie D X, Sun C Q. PAY1 improves plant architecture and enhances grain yield in rice[J]. Plant Journal, 2015, 83(3): 528-536. |
| [23] | Harmoko R, Yoo J Y, Ko K S, Ramasamy N K, Hwang B Y, Lee E J, Kim H S, Lee K J, Oh D B, Kim D Y, Lee S H, Li Y, Lee S Y, Lee K O. N-glycan containing a core α1, 3-fucose residue is required for basipetal auxin transport and gravitropic response in rice (Oryza sativa)[J]. New Phytologist, 2016, 212(1): 108-122. |
| [24] | Wang W S, Mauleon R, Hu Z Q, Chebotarov D, Tai S S, Wu Z C, Li M, Zheng T Q, Fuentes R R, Zhang F, Mansueto L, Li Z K, Leung H. Genomic variation in 3,010 diverse accessions of Asian cultivated rice[J]. Nature, 2018, 557(7703): 43-49. |
| [25] | 沈圣泉, 庄杰云, 包劲松, 郑康乐, 夏英武, 舒庆尧. 水稻分蘖最大角度的QTL分析[J]. 农业生物技术学报, 2005(1): 16-20. |
| Shen S Q, Zhuang J Y, Bao J S, Zheng K L, Xia Y W, Shu Q Y. QTL analysis of maximum tiller angle in rice[J]. Journal of Agricultural Biotechnology, 2005(1): 16-20. (in Chinese with English abstract) | |
| [26] | Bradbury P J, Zhang Z, Kroon D E, Casstevens T M, Ramdoss Y, Buckler E S. TASSEL: Software for association mapping of complex traits in diverse samples[J]. Bioinformatics, 2007, 23(19): 2633-2635. |
| [27] | Zhao K, Tung C W, Eizenga G C, Wright M H, Ali M L, Price A H, Norton G J, Islam M R, Reynolds A, Mezey J, McClung A M, Bustamante C D, McCouch S R. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa[J]. Nature Communications, 2011, 2: 467. |
| [28] | Bai S H, Hong J, Su S, Li Z K, Wang W S, Shi J X, Liang W Q, Zhang D B. Genetic basis underlying tiller angle in rice by genome-wide association study[J]. Plant Cell Reports, 2022, 41(8): 1707-1720. |
| [29] | 梁彦, 王永红. 水稻株型功能基因及其在育种上的应用[J]. 生命科学, 2016, 28(10): 1156-1167. |
| Liang Y, Wang Y H. The genes controlling rice architecture and its application in breeding[J]. Chinese Bulletin: Life Science, 2016, 28(10): 1156-1167. (in Chinese with English abstract) | |
| [30] | 陈宗祥, 冯志明, 王龙平, 冯凡, 张亚芳, 马玉银, 潘学彪, 左示敏. 水稻分蘖角基因TAC1的育种应用价值分析[J]. 中国水稻科学, 2017, 31(6): 590-598. |
| Chen Z X, Feng Z M, Wang L P, Feng F, Zhang Y F, Ma Y Y, Pan X B, Zuo S M. Breeding potential of rice TAC1 gene for tiller angle[J]. Chinese Journal of Rice Science, 2017, 31(6): 590-598. (in Chinese with English abstract) | |
| [31] | 李珍珠, 彭清祥, 邱先进, 徐俊英, 李志新, 刘海洋. 水稻分蘖角度基因TIG1功能性分子标记的开发和应用[J]. 植物遗传资源学报, 2023, 24(3): 808-816. |
| Li Z Z, Peng Q X, Qiu X J, Xu J Y, Li Z X, Liu H Y, Development and application of functional molecular marker of rice tiller angle gene TIG1[J]. Journal of Plant Genetic Resources, 2023, 24(3): 808-816. (in Chinese with English abstract) | |
| [32] | Wang L, Xu Y Y, Zhang C, Ma Q B, Joo S H, Kim S K, Xu Z H, Chong K. OsLIC, a novel CCCH-Type zinc finger protein with transcription activationmediates rice architecture via brassinosteroids signaling[J]. PLoS One, 2008, 3(10): e3521. |
| [1] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [2] | LIU Zhongqi, ZHANG Haiqing, HE Jiwai, GUI Jinxin. Genome-wide Association Analysis of Rice Seed Dehydration Rate at Maturity Stage [J]. Chinese Journal OF Rice Science, 2024, 38(2): 150-159. |
| [3] | HOU Benfu, YANG Chuanming, ZHANG Xijuan, YANG Xianli, WANG Lizhi, WANG Jiayu, LI Hongyu, JIANG Shukun. Mapping of Grain Shape QTLs Using RIL Population from Longdao 5/Zhongyouzao 8 [J]. Chinese Journal OF Rice Science, 2024, 38(1): 13-24. |
| [4] | HU Jiaxiao, LIU Jin, CUI Di, LE Si, ZHOU Huiying, HAN Bing, MENG Bingxin, YU Liqin, HAN Longzhi, MA Xiaoding, LI Maomao. Mapping Major QTLs for Panicle Traits Using CSSLs of Dongxiang Wild Rice (Oryza rufipogon Griff.) [J]. Chinese Journal OF Rice Science, 2023, 37(6): 597-608. |
| [5] | XIE Kaizhen, ZHANG Jianming, CHENG Can, ZHOU Jihua, NIU Fuan, SUN Bin, ZHANG Anpeng, WEN Weijun, DAI Yuting, HU Qiyan, QIU Yue, CAO Liming, CHU Huangwei. Identification and QTL Mapping of Rice Germplasm Resources with Low Amylose Content [J]. Chinese Journal OF Rice Science, 2023, 37(6): 609-616. |
| [6] | YAO Xiaoyun, CHEN Chunlian, XIONG Yunhua, HUANG Yongping, PENG Zhiqing, LIU Jin, YIN Jianhua. Identification of QTL for Milling and Appearance Quality Traits in Rice (Oryza sativa L.) [J]. Chinese Journal OF Rice Science, 2023, 37(5): 507-517. |
| [7] | WEI Minyi, MA Zengfeng, HUANG Dahui, QIN Yuanyuan, LIU Chi, LU Yingping, LUO Tongping, LI Zhenjing, ZHANG Yuexiong, QIN Gang. QTL-Seq Analysis for Identification of Resistance Locus to Bacterial Leaf Streak in Rice [J]. Chinese Journal OF Rice Science, 2023, 37(2): 133-141. |
| [8] | LIU Jin, CUI Di, YU Liqin, ZHANG Lina, ZHOU Huiying, MA Xiaoding, HU Jiaxiao, HAN Bing, HAN Longzhi, LI Maomao. Screening and QTL Mapping of Heat-tolerant Rice (Oryza sativa L.) Germplasm Resources at Seedling Stage [J]. Chinese Journal OF Rice Science, 2022, 36(3): 259-268. |
| [9] | HUANG Tao, WANG Yanning, ZHONG Qi, CHENG Qin, YANG Mengmeng, WANG Peng, WU Guangliang, HUANG Shiying, LI Caijing, YU Jianfeng, HE Haohua, BIAN Jianmin. Mapping and Analysis of QTLs for Rice Grain Weight and Grain Shape Using Chromosome Segment Substitution Line Population [J]. Chinese Journal OF Rice Science, 2022, 36(2): 159-170. |
| [10] | DONG Zheng, WANG Yamei, LI Yongchao, XIONG Haibo, XUE Canhui, PAN Xiaowu, LIU Wenqiang, WEI Xiucai, LI Xiaoxiang. Genome-wide Association Analysis of Cadmium Content in Rice Based on MAGIC Population [J]. Chinese Journal OF Rice Science, 2022, 36(1): 35-42. |
| [11] | Jie LI, Rongrong TIAN, Tianliang BAI, Chunyan ZHU, Jiawei SONG, Lei TIAN, Shuaiguo MA, Jiandong LÜ, Hui HU, Zhenyu WANG, Chengke LUO, Yinxia ZHANG, Peifu LI. Comprehensive Evaluation and QTL Analysis for Flag Leaf Traits Using a Backcross Population in Rice [J]. Chinese Journal OF Rice Science, 2021, 35(6): 573-585. |
| [12] | Xina CHEN, Zeke YUAN, Zhenzhen HU, Quanzhi ZHAO, Hongzheng SUN. QTL-Seq Mapping of Head Rice Rate QTLs in japonica Rice [J]. Chinese Journal OF Rice Science, 2021, 35(5): 449-454. |
| [13] | Chengxing DU, Huali ZHANG, Dongqing DAI, Mingyue WU, Minmin LIANG, Junyu CHEN, Liangyong MA. QTL Analysis for Grain Weight and Shape and Validation of qTGW1.2/qGL1.2 [J]. Chinese Journal OF Rice Science, 2021, 35(4): 359-372. |
| [14] | Yujun ZHU, Ziwei ZUO, Zhenhua ZHANG, Yeyang FAN. A New Approach for Fine-mapping and Map-based Cloning of Minor-Effect QTL in Rice [J]. Chinese Journal OF Rice Science, 2021, 35(4): 407-414. |
| [15] | Wei LIU, Zhanhua LU, Dongbai LU, Xiaofei WANG, Shiguang WANG, Jia XUE, Xiuying HE. Location and Candidate Gene Analysis of Rice Clustered Spikelets Gene OsCL6 [J]. Chinese Journal OF Rice Science, 2021, 35(3): 238-248. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||