
Chinese Journal OF Rice Science ›› 2024, Vol. 38 ›› Issue (2): 150-159.DOI: 10.16819/j.1001-7216.2024.230305
• Research Papers • Previous Articles Next Articles
LIU Zhongqi1,2, ZHANG Haiqing1, HE Jiwai1,*( ), GUI Jinxin1
), GUI Jinxin1
Received:2023-03-17
Revised:2023-06-16
Online:2024-03-10
Published:2024-03-14
Contact:
* email: hejiwai@hunau.edu.cn
通讯作者:
* email: hejiwai@hunau.edu.cn
基金资助:LIU Zhongqi, ZHANG Haiqing, HE Jiwai, GUI Jinxin. Genome-wide Association Analysis of Rice Seed Dehydration Rate at Maturity Stage[J]. Chinese Journal OF Rice Science, 2024, 38(2): 150-159.
刘忠奇, 张海清, 贺记外, 桂金鑫. 成熟期水稻种子脱水速率全基因组关联分析[J]. 中国水稻科学, 2024, 38(2): 150-159.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2024.230305
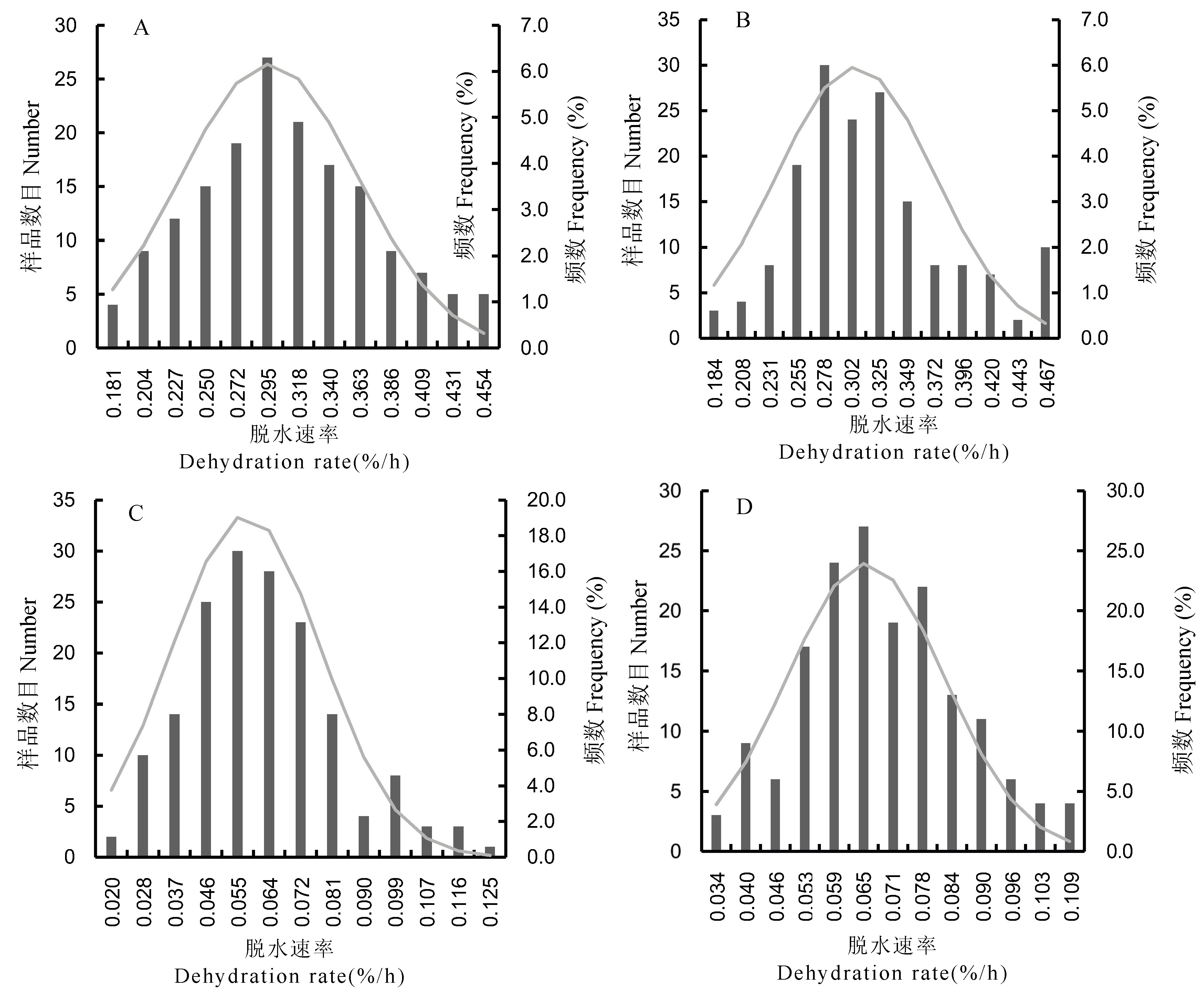
Fig. 1. Distribution of rice seed dehydration rates of tested rice material A, Rapid dehydration treatment(2019); B, Rapid dehydration treatment (2020); C, Slow dehydration treatment(2019); D, Slow dehydration treatment(2020). The fast dehydration condition is constant temperature 45 ± 2℃, the slow dehydration condition is constant temperature 40 ± 2℃.
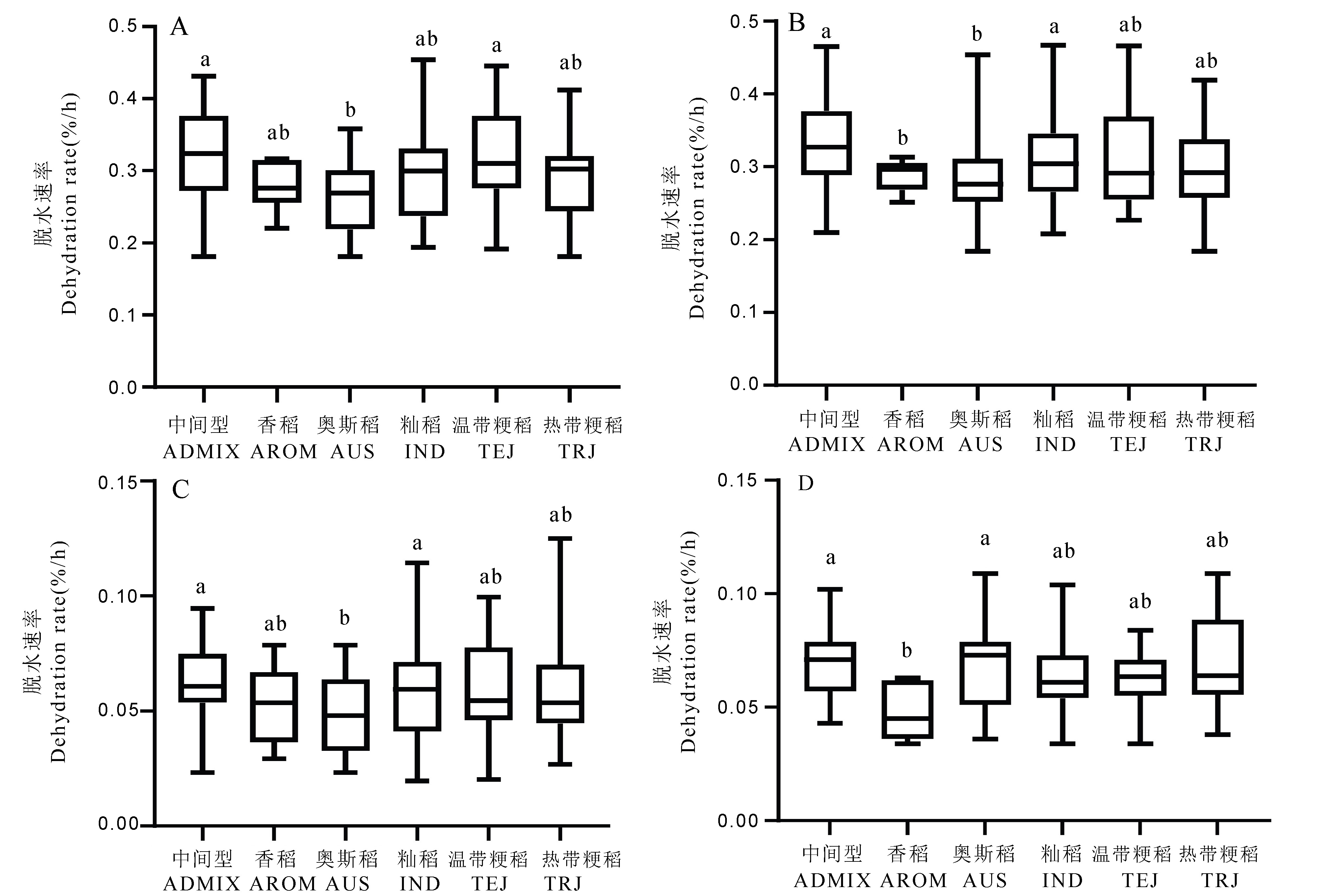
Fig. 2. Box plots of dehydration rate of different sub-groups of rice under different dehydration conditions in two years A, Rapid dehydration treatment(2019); B, Rapid dehydration treatment(2020); C, Slow dehydration treatment(2019); D, Slow dehydration treatment(2020). Different lowercase letters indicate significant difference of 0.05 level among different subgroups (Duncan multiple comparison). ADMIX, Intermediate type; AROM, Aromatic rice; AUS, Aus rice; IND, indica rice; TEJ, Temperate japonica rice; TRJ, Tropical japonica rice.

Fig. 3. Manhattan map and QQ map of QTLs for seed dehydration rate at maturity under different dehydration treatments A, Manhattan plot of QTLs for seed dehydration rate at maturity under the fast dehydration conditions in 2019. The areas above the red solid line represent significant difference at P < 0.001 level, and the same below; B, QQ map of QTLs for seed dehydration rate at maturity under the fast dehydration conditions in 2019; C and D, Under the fast dehydration conditions in 2020; E and F, Under the low dehydration conditions in 2019; G and H, Under the low dehydration conditions in 2020.
| 染色体Chromosome | 位点 Locus | 候选基因 Candidate gene | 功能注释 Functional annotation | 参考基因 Reference gene | |
|---|---|---|---|---|---|
| 1 | qGDR1.2 | LOC_Os01g49710 | 谷胱甘肽S-转移酶 Glutathione S-transferase | ||
| 2 | qGDR2.2 | LOC_Os02g36974 | 14-3-3蛋白 14-3-3 protein | ||
| 2 | qGDR2.3 | LOC_Os02g44630 | 水通道蛋白 Aquaporin protein | OsPIP1;1[ | |
| 4 | qGDR4.1 | LOC_Os04g32480 | 锌指蛋白 Zinc-finger protein | OsTIFY9[ | |
| 4 | qGDR4.2 | LOC_Os04g53240 | 自噬相关蛋白 Autophagy-related protein | OsATG8b[ | |
| 6 | qGDR6.1 | LOC_Os06g03670 | 脱水反应元件结合蛋白 Dehydration-responsive element-binding protein | OsDREB1C[ | |
| 6 | qGDR6.2 | LOC_Os06g12350 | 氨基酸转运蛋白 Amino acid transporter | ||
| 6 | qGDR6.4 | LOC_Os06g39960 | bZIP转录因子结构域含蛋白 bZIP transcription factor domain containing protein | OsbZIP48[ | |
| 7 | qGDR7.3 | LOC_Os07g37890 | 蛋白磷酸酶2C Protein phosphatase 2C | ||
| 7 | qGDR7.4 | LOC_Os07g38580 | 锌指家族蛋白 Zinc finger family protein | ||
| 7 | qGDR7.6 | LOC_Os07g47250 | 脂肪酶前体 Lipase precursor | ||
| 8 | qGDR8.2 | LOC_Os08g06240 | MYB家族转录因子 MYB family transcription factor | ||
| 8 | qGDR8.3 | LOC_Os08g15149 | 硫氧还蛋白结构域蛋白9 Thioredoxin domain-containing protein 9 | ||
| 9 | qGDR9.1 | LOC_Os09g20990 | 海藻糖-6-磷酸合成酶 Trehalose-6-phosphate synthase | ||
| 9 | qGDR9.2 | LOC_Os09g26170 | MYB家族转录因子 MYB family transcription factor | ||
| 10 | qGDR10.1 | LOC_Os10g01134 | OsSCP46-推定丝氨酸羧肽酶同源物 OsSCP46-putative Serine Carboxypeptidase homologue | OsSCP46[ | |
| 10 | qGDR10.3 | LOC_Os10g25010 | OsCML8-钙调素相关钙传感器蛋白 OsCML8-calmodulin-related calcium sensor protein | ||
| 11 | qGDR11.1 | LOC_Os11g37000 | 热激蛋白DnaJ Heat shock protein DnaJ | ||
Table 1. Candidate genes and their functional annotations for predicting the rate of seed dehydration in maturity
| 染色体Chromosome | 位点 Locus | 候选基因 Candidate gene | 功能注释 Functional annotation | 参考基因 Reference gene | |
|---|---|---|---|---|---|
| 1 | qGDR1.2 | LOC_Os01g49710 | 谷胱甘肽S-转移酶 Glutathione S-transferase | ||
| 2 | qGDR2.2 | LOC_Os02g36974 | 14-3-3蛋白 14-3-3 protein | ||
| 2 | qGDR2.3 | LOC_Os02g44630 | 水通道蛋白 Aquaporin protein | OsPIP1;1[ | |
| 4 | qGDR4.1 | LOC_Os04g32480 | 锌指蛋白 Zinc-finger protein | OsTIFY9[ | |
| 4 | qGDR4.2 | LOC_Os04g53240 | 自噬相关蛋白 Autophagy-related protein | OsATG8b[ | |
| 6 | qGDR6.1 | LOC_Os06g03670 | 脱水反应元件结合蛋白 Dehydration-responsive element-binding protein | OsDREB1C[ | |
| 6 | qGDR6.2 | LOC_Os06g12350 | 氨基酸转运蛋白 Amino acid transporter | ||
| 6 | qGDR6.4 | LOC_Os06g39960 | bZIP转录因子结构域含蛋白 bZIP transcription factor domain containing protein | OsbZIP48[ | |
| 7 | qGDR7.3 | LOC_Os07g37890 | 蛋白磷酸酶2C Protein phosphatase 2C | ||
| 7 | qGDR7.4 | LOC_Os07g38580 | 锌指家族蛋白 Zinc finger family protein | ||
| 7 | qGDR7.6 | LOC_Os07g47250 | 脂肪酶前体 Lipase precursor | ||
| 8 | qGDR8.2 | LOC_Os08g06240 | MYB家族转录因子 MYB family transcription factor | ||
| 8 | qGDR8.3 | LOC_Os08g15149 | 硫氧还蛋白结构域蛋白9 Thioredoxin domain-containing protein 9 | ||
| 9 | qGDR9.1 | LOC_Os09g20990 | 海藻糖-6-磷酸合成酶 Trehalose-6-phosphate synthase | ||
| 9 | qGDR9.2 | LOC_Os09g26170 | MYB家族转录因子 MYB family transcription factor | ||
| 10 | qGDR10.1 | LOC_Os10g01134 | OsSCP46-推定丝氨酸羧肽酶同源物 OsSCP46-putative Serine Carboxypeptidase homologue | OsSCP46[ | |
| 10 | qGDR10.3 | LOC_Os10g25010 | OsCML8-钙调素相关钙传感器蛋白 OsCML8-calmodulin-related calcium sensor protein | ||
| 11 | qGDR11.1 | LOC_Os11g37000 | 热激蛋白DnaJ Heat shock protein DnaJ | ||
| [1] | 刘忠奇, 贺记外, 张海清, 刘爱民. 植物种子脱水耐性的研究现状分析与展望[J]. 中国农学通报, 2020, 36(2): 36-41. |
| Liu Z Q, He J W, Zhang H Q, Liu A M. Dehydration tolerance of plant seeds: Current research situation and prospects[J]. Chinese Agricultural Science Bulletin, 2020, 36(2): 36-41. (in Chinese with English abstract) | |
| [2] | Zhou G F, Hao D R, Xue L, Chen G Q, Lu H H, Zhang Z L, Shi M L, Mao Y X. Genome-wide association study of kernel moisture content at harvest stage in maize[J]. Breeding Science, 2018, 68(5): 622-628. |
| [3] | Jia T J, Wang L F, Li J J, Ma J, Cao Y Y, Lübbersted T, Li H Y. Integrating a genome-wide association study with transcriptomic analysis to detect genes controlling grain drying rate in maize (Zea mays L)[J]. Theoretical and Applied Genetics, 2020, 133(2): 623-634. |
| [4] | 刘显君, 王振华, 王霞, 李庭锋, 张林. 玉米籽粒生理成熟后自然脱水速率QTL的初步定位[J]. 作物学报, 2010, 36(1): 47-52. |
| Liu X J, Wang Z H, Wang X, Li T F, Zhang L. Primary mapping of QTL for dehydration rate of maize kernel after physiological maturing[J]. Acta Agronomica Sinica, 2010, 36(1): 47-52. (in Chinese with English abstract) | |
| [5] | Sala R G, Andrade F H, Camadro E L, Cerono J C. Quantitative trait loci for grain moisture at harvest and field grain drying rate in maize (Zea mays L.)[J]. Theoretical and Applied Genetics, 2006, 112: 462-471. |
| [6] | Li Y L, Dong Y B, Yang M L, Wang Q L, Shi Q, Zhou Q, Deng F, Ma Z Y, Qiao D H, Xu H. QTL detection for grain water relations and genetic correlations with grain matter accumulation at four stages after pollination in maize[J]. Journal of Plant Biochemistry and Physiology, 2014, 2(1): 1-9. |
| [7] | Capelle V, Remoué C, Moreau L, Reyss A, MahéahssMassonneau A, Falque M, Charcosset A, Thévenot C, Rogowsky P, Coursol S, Prioul J L. QTLs and candidate genes for desiccation and abscisic acid content in maize kernels[J]. BMC Plant Biology, 2010, 10(1): 1-22. |
| [8] | 孙乐秀. 玉米自交系机械化收获相关性状及其SNP标记关联分析[D]. 泰安: 山东农业大学, 2015. |
| Sun L X. Traits related to mechanical harvesting and their association analysis with SNP markers in maize inbred lines[D]. Tai’an: Shandong Agricultural University, 2015. (in Chinese with English abstract) | |
| [9] | Li W Q, Yu Y H, Wang L X, Luo Y, Peng Y, Xu Y C, Liu X G, Wu S S, Jian L M, Xu J T, Xiao Y J, Yan J B. The genetic architecture of the dynamic changes in grain moisture in maize[J]. Plant Biotechnology Journal, 2021, 19: 1195-1205. |
| [10] | 朱冬梅, 胡文静, 别同德, 陆成彬, 赵仁慧, 高德荣. 利用四交RIL群体定位小麦籽粒脱水速率 QTL[J]. 麦类作物学报, 2020, 40(1): 49-54. |
| Zhu D M, Hu W J, Bie T D, Lu C B, Zhao R H, Gao D R. QTL mapping for kernel dehydration rate after physiological maturity using in four way RIL populations of wheat[J]. Journal of Triticeae Crop, 2020, 40(1): 49-54. (in Chinese with English abstract) | |
| [11] | 于澎湃. 高粱耐密性研究及籽粒脱水速率的QTL定位[D]. 天津: 天津农学院, 2019. |
| Yu P P. Studies on sorghum density tolerance and QTL mapping of grain dehydration rate[D]. Tianjin:Tianjin Agricultural University, 2019. (in Chinese with English abstract) | |
| [12] | 袁静, 任育军, 赵洁. 水稻胚胎发育过程相关基因的鉴定[J]. 分子细胞生物学报, 2008, 41(3): 238-244. |
| Yuan J, Ren Y J, Zhao J. Identification of genes associated with early and late embryo development in rice[J]. Journal of Molecular Cell Biology, 2008, 41(3): 238-244. (in Chinese with English abstract) | |
| [13] | 张娟, 牛百晓, 鄂志国, 陈忱. 水稻胚乳发育遗传调控的研究进展[J]. 中国水稻科学, 2021, 35(4): 326-341. |
| Zhang J, Niu B X, E Z G, Chen C. Towards understanding the genetic regulations of endosperm development in rice[J]. Chinese Journal of Rice Science, 2021, 35(4): 326-341. (in Chinese with English abstract) | |
| [14] | Samarah N H, Mullen R E, Alqudah A M. An index to quantify seed moisture loss rate in relationship with seed desiccation tolerance in common vetch[J]. Seed Science and Technology, 2009, 37: 413-422. |
| [15] | Zhao K Y, Tung C W, Eizenga G C, Wright M H, Ali M L, Price A H, Norton G J, Islam M R, Reynolds A, Mezey J, McClung A M, Bustamante C D, McCouch S R. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa[J]. Nature Communications, 2011, 2: 467-477. |
| [16] | 林晋军, 姚威, 彭沙莎, 刘文韬, 陈文凤, 刘雄伦, 刘金灵. 水稻抽穗期性状QTL的全基因组关联定位分析[J]. 湖南农业大学学报, 2021, 47(3): 283-290. |
| Lin J J, Yao W, Peng S S, Liu W T, Chen W F, Liu X L, Liu J L. Genome-wide association dissection of QTLs for heading date traits in rice[J]. Journal of Hunan Agricultural University, 2021, 47(3): 283-290. (in Chinese with English abstract) | |
| [17] | Liu C W, Tatsuya F, Tadashi M, Gena P, Frascaria D, Kaneko T, Katsuhara M, Zhong S H, Sun X L, Zhu Y M, Iwasaki I, Kitagawa Y L. Aquaporin OsPIP1;1 promotes rice salt resistance and seed germination[J]. Plant Physiology and Biochemistry, 2013, 63: 151-158. |
| [18] | Dubouzet J G, Sakuma Y, Ito Y, Kasuga M, Dubouzet E G, Miura S, Seki M, Shinozak K, Shinozaki Y. OsDREB genes in rice, encode transcription activators that function in drought, high-salt and cold responsive gene expression[J]. The Plant Journal, 2003, 33(4): 751-763. |
| [19] | Li Z Y, Tang L Q, Qiu J H, Zhang W, Wang Y F, Tong X H, Wei X J, Hou Y X, Zhang J. Serine carboxypeptidase 46 regulates grain filling and seed germination in rice[J]. PLoS One, 2016, 11(7): 1-18. |
| [20] | Ji Q, Zhang L S, Wang Y F, Wang J. Genome-wide analysis of basic leucine zipper transcription factor families in Arabidopsis thaliana, Oryza sativa and Populus trichocarpa[J]. Journal of Shanghai University, 2009, 13(2): 174-182. |
| [21] | Ye H, Du H, Tang N, Li X, Xiong L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice[J]. Plant Molecular Biology, 2009, 71(3): 291-305. |
| [22] | Izumi M, Hidema J, Wada S, Kondo E, Makino A, Ishida H. Establishment of monitoring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation[J]. Plant Physiology, 2015, 167(4): 1307-1320. |
| [23] | 渠建洲. 玉米籽粒含水率和脱水速率的基因挖掘与分析[D]. 杨凌: 西北农林科技大学, 2021. |
| Qu J Z. Gene mining and analysis of maize kernel moisture content and dehydration rate[D]. Yangling: Northwest A&F University, 2021. (in Chinese with English abstract) | |
| [24] | Si J K, Arifa A. Tissue and cell-specific localization of rice aquaporins and their water transport activities[J]. Plant and Cell Physiology, 2008, 49(1): 30-39. |
| [25] | 张凤启, 王邑双, 丁勇, 张君, 赵霞, 赵发欣, 唐保军. 玉米籽粒脱水速率研究进展[J]. 农学学报, 2018, 8(11): 4-11. |
| Zhang F Q, Wang Y S, Ding Y, Zhang J, Zhao X, Zhao F X, Tang B J. Corn kernel dehydration rate: Research progress[J]. Journal of Agriculture, 2018, 8(11): 4-11. (in Chinese with English abstract) | |
| [26] | Peng B, Kong H L, Li Y B, Wang L Q, Zhong M, Sun L, Gao G J, Zhang Q L, Luo L J, Wang G W, Xie W B, Chen J X, Yao W, Peng Y, Lei L, Lian X M, Xiao J H, Xu C G, Li X H, He Y Q. OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice[J]. Nature Communications, 2014, 5: 5847. (in Chinese with English abstract) |
| [27] | 彭波, 孙艳芳, 庞瑞华, 周棋赢, 袁红雨, 宋世枝. 植物氨基酸转运蛋白的研究进展[J]. 热带作物学报, 2016, 37(6): 1238-1243. |
| Peng B, Sun Y F, Pang R H, Zhou Q Y, Yuan H Y, Song S Z. Research progress of amino acid transporters in plants[J]. Chinese Journal of Tropical Crops, 2016, 37(6): 1238-1243. (in Chinese with English abstract) | |
| [28] | 叶玲飞, 罗光宇, 向建华, 刘爱玲, 陈信波. 丝氨酸羧肽酶及其类蛋白的研究进展[J]. 植物生理学报, 2013, 49(2): 122-126. |
| Ye L F, Luo G Y, Xiang J H, Liu A L, Chen X B. Research progress of serine carboxypeptidases and serine carboxypeptidase like proteins[J]. Plant Physiology Journal, 2013, 49(2): 122-126. (in Chinese with English abstract) | |
| [29] | 叶玲飞. 水稻OsSCP基因的耐逆相关功能研究[D]. 长沙: 湖南农业大学, 2013. |
| Ye L F. Studies on the stress resistance functions of OsSCP gene[D]. Changsha: Hunan Agricultural University, 2013. (in Chinese with English abstract) |
| [1] |
LÜ Yang, LIU Congcong, YANG Longbo, CAO Xinglan, WANG Yueying, TONG Yi, Mohamed Hazman, QIAN Qian, SHANG Lianguang, GUO Longbiao.
Identification of Candidate Genes for Rice Nitrogen Use Efficiency by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(5): 516-524. |
| [2] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [3] | ZHENG Guangjie, YE Chang, ZHU Junlin, TAO Yi, XIAO Deshun, XU Yanan, CHU Guang, XU Chunmei, WANG Danying. Relationship Between Embryo Survival and Glucose Supply of Rice Seed and Embryo Under Flooding Stress [J]. Chinese Journal OF Rice Science, 2024, 38(2): 172-184. |
| [4] | HOU Benfu, YANG Chuanming, ZHANG Xijuan, YANG Xianli, WANG Lizhi, WANG Jiayu, LI Hongyu, JIANG Shukun. Mapping of Grain Shape QTLs Using RIL Population from Longdao 5/Zhongyouzao 8 [J]. Chinese Journal OF Rice Science, 2024, 38(1): 13-24. |
| [5] | HU Jiaxiao, LIU Jin, CUI Di, LE Si, ZHOU Huiying, HAN Bing, MENG Bingxin, YU Liqin, HAN Longzhi, MA Xiaoding, LI Maomao. Mapping Major QTLs for Panicle Traits Using CSSLs of Dongxiang Wild Rice (Oryza rufipogon Griff.) [J]. Chinese Journal OF Rice Science, 2023, 37(6): 597-608. |
| [6] | XIE Kaizhen, ZHANG Jianming, CHENG Can, ZHOU Jihua, NIU Fuan, SUN Bin, ZHANG Anpeng, WEN Weijun, DAI Yuting, HU Qiyan, QIU Yue, CAO Liming, CHU Huangwei. Identification and QTL Mapping of Rice Germplasm Resources with Low Amylose Content [J]. Chinese Journal OF Rice Science, 2023, 37(6): 609-616. |
| [7] | YAO Xiaoyun, CHEN Chunlian, XIONG Yunhua, HUANG Yongping, PENG Zhiqing, LIU Jin, YIN Jianhua. Identification of QTL for Milling and Appearance Quality Traits in Rice (Oryza sativa L.) [J]. Chinese Journal OF Rice Science, 2023, 37(5): 507-517. |
| [8] | WEI Minyi, MA Zengfeng, HUANG Dahui, QIN Yuanyuan, LIU Chi, LU Yingping, LUO Tongping, LI Zhenjing, ZHANG Yuexiong, QIN Gang. QTL-Seq Analysis for Identification of Resistance Locus to Bacterial Leaf Streak in Rice [J]. Chinese Journal OF Rice Science, 2023, 37(2): 133-141. |
| [9] | LIU Jin, CUI Di, YU Liqin, ZHANG Lina, ZHOU Huiying, MA Xiaoding, HU Jiaxiao, HAN Bing, HAN Longzhi, LI Maomao. Screening and QTL Mapping of Heat-tolerant Rice (Oryza sativa L.) Germplasm Resources at Seedling Stage [J]. Chinese Journal OF Rice Science, 2022, 36(3): 259-268. |
| [10] | HUANG Tao, WANG Yanning, ZHONG Qi, CHENG Qin, YANG Mengmeng, WANG Peng, WU Guangliang, HUANG Shiying, LI Caijing, YU Jianfeng, HE Haohua, BIAN Jianmin. Mapping and Analysis of QTLs for Rice Grain Weight and Grain Shape Using Chromosome Segment Substitution Line Population [J]. Chinese Journal OF Rice Science, 2022, 36(2): 159-170. |
| [11] | DONG Zheng, WANG Yamei, LI Yongchao, XIONG Haibo, XUE Canhui, PAN Xiaowu, LIU Wenqiang, WEI Xiucai, LI Xiaoxiang. Genome-wide Association Analysis of Cadmium Content in Rice Based on MAGIC Population [J]. Chinese Journal OF Rice Science, 2022, 36(1): 35-42. |
| [12] | Jie LI, Rongrong TIAN, Tianliang BAI, Chunyan ZHU, Jiawei SONG, Lei TIAN, Shuaiguo MA, Jiandong LÜ, Hui HU, Zhenyu WANG, Chengke LUO, Yinxia ZHANG, Peifu LI. Comprehensive Evaluation and QTL Analysis for Flag Leaf Traits Using a Backcross Population in Rice [J]. Chinese Journal OF Rice Science, 2021, 35(6): 573-585. |
| [13] | Xina CHEN, Zeke YUAN, Zhenzhen HU, Quanzhi ZHAO, Hongzheng SUN. QTL-Seq Mapping of Head Rice Rate QTLs in japonica Rice [J]. Chinese Journal OF Rice Science, 2021, 35(5): 449-454. |
| [14] | Chengxing DU, Huali ZHANG, Dongqing DAI, Mingyue WU, Minmin LIANG, Junyu CHEN, Liangyong MA. QTL Analysis for Grain Weight and Shape and Validation of qTGW1.2/qGL1.2 [J]. Chinese Journal OF Rice Science, 2021, 35(4): 359-372. |
| [15] | Yujun ZHU, Ziwei ZUO, Zhenhua ZHANG, Yeyang FAN. A New Approach for Fine-mapping and Map-based Cloning of Minor-Effect QTL in Rice [J]. Chinese Journal OF Rice Science, 2021, 35(4): 407-414. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||