
Chinese Journal OF Rice Science ›› 2023, Vol. 37 ›› Issue (5): 478-485.DOI: 10.16819/j.1001-7216.2023.220907
• Research Papers • Previous Articles Next Articles
LI Jingfang1, WEN Shuyue2, ZHAO Lijun2, CHEN Tingmu1, ZHOU Zhenling1, SUN Zhiguang1, LIU Yan1, CHEN Haiyuan3, ZHANG Yunhui3, CHI Ming1, XING Yungao1, XU Bo1, XU Dayong1, WANG Baoxiang1,*( )
)
Received:2022-09-28
Revised:2022-12-19
Online:2023-09-10
Published:2023-09-13
Contact:
*email:
李景芳1, 温舒越2, 赵利君2, 陈庭木1, 周振玲1, 孙志广1, 刘艳1, 陈海元3, 张云辉3, 迟铭1, 邢运高1, 徐波1, 徐大勇1, 王宝祥1,*( )
)
通讯作者:
*email:基金资助:LI Jingfang, WEN Shuyue, ZHAO Lijun, CHEN Tingmu, ZHOU Zhenling, SUN Zhiguang, LIU Yan, CHEN Haiyuan, ZHANG Yunhui, CHI Ming, XING Yungao, XU Bo, XU Dayong, WANG Baoxiang. Development of Aromatic Salt-tolerant Rice Based on CRISPR/Cas9 Technology[J]. Chinese Journal OF Rice Science, 2023, 37(5): 478-485.
李景芳, 温舒越, 赵利君, 陈庭木, 周振玲, 孙志广, 刘艳, 陈海元, 张云辉, 迟铭, 邢运高, 徐波, 徐大勇, 王宝祥. 基于CRISPR/Cas9技术创制耐盐香稻[J]. 中国水稻科学, 2023, 37(5): 478-485.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2023.220907
| 引物名称 Primer name | 引物序列 Primer sequence(5’-3’) | 用途 Usage | |
|---|---|---|---|
| Badh2-S1-F | GGCATGGTGGAAAAAGTCCTATAG | 构建Badh2敲除载体 Construction of Badh2 knockout vector | |
| Badh2-S1-R | AAACCTATAGGACTTTTTCCACCA | ||
| Badh2-F | CCTTTGTCATCACACCCTGG | Badh2敲除靶点测序鉴定 | |
| Badh2-R | TAACTGCCTTCCTTGCCACG | Sequencing for Badh2 knockout target | |
| OsRR22-S1-F | GGCACTCAACTGACCATACAACTC | 构建OsRR22敲除载体 | |
| OsRR22-S1-R | AAACGAGTTGTATGGTCAGTTGAG | Construction of OsRR22 knockout vector | |
| OsRR22-F | TAGGAGGAAGTTCGGTAATCGT | OsRR22敲除靶点测序鉴定 | |
| OsRR22-R | ACATTTTCCCTGGTGAGTTTCT | Sequencing for OsRR22 knockout target | |
| Cas9-F | CCTTCTGTGTGGTCTGGTTC | 载体检测 | |
| Cas9-R | AGACAATCACCCCCTGGAAC | Vector assay | |
| Hyg-F | CTATTTCTTTGCCCTCGGAC | 转基因检测 | |
| Hyg-R | ATGCCTGAACTCACCGCGAC | Transgenic detection | |
| qPCR SOS1-F | ATTGCAGCTGAGCATGTACG | 实时荧光定量PCR | |
| qPCR SOS1-R | AGAGCTTGCTTTCGTGTGAC | Quantitative real-time PCR | |
| qPCR OsMYB2-F | GGGCTGAAACGCACAGGCAAGA | 实时荧光定量PCR | |
| qPCR OsMYB2-R | CTGCTTGGCGTGCTTCTGC | Quantitative real-time PCR | |
| qPCR OsNHX1-F | GTGACAGACCTGGCAAATCC | 实时荧光定量PCR | |
| qPCR OsNHX1-R | TCGACACAGCTCCTCTCATC | Quantitative real-time PCR | |
| qPCR OsHKT1;1-F | TGCCAGAAGTTGTTGAAGCC | 实时荧光定量PCR | |
| qPCR OsHKT1;1-R | CCCAGGAACATCACCAGGAT | Quantitative real-time PCR | |
| qPCR OsP5CS1-F | GTCAGAGTGGACTGATGGCT | 实时荧光定量PCR | |
| qPCR OsP5CS1-R | GCCTTTCTAGTGCTGATGGC | Quantitative real-time PCR | |
| UBIQUITIN-F | GCTCCGTGGCGGTATCAT | 实时荧光定量PCR | |
| UBIQUITIN-R | CGGCAGTTGACAGCCCTAG | Quantitative real-time PCR | |
Table 1. Primers used in this study.
| 引物名称 Primer name | 引物序列 Primer sequence(5’-3’) | 用途 Usage | |
|---|---|---|---|
| Badh2-S1-F | GGCATGGTGGAAAAAGTCCTATAG | 构建Badh2敲除载体 Construction of Badh2 knockout vector | |
| Badh2-S1-R | AAACCTATAGGACTTTTTCCACCA | ||
| Badh2-F | CCTTTGTCATCACACCCTGG | Badh2敲除靶点测序鉴定 | |
| Badh2-R | TAACTGCCTTCCTTGCCACG | Sequencing for Badh2 knockout target | |
| OsRR22-S1-F | GGCACTCAACTGACCATACAACTC | 构建OsRR22敲除载体 | |
| OsRR22-S1-R | AAACGAGTTGTATGGTCAGTTGAG | Construction of OsRR22 knockout vector | |
| OsRR22-F | TAGGAGGAAGTTCGGTAATCGT | OsRR22敲除靶点测序鉴定 | |
| OsRR22-R | ACATTTTCCCTGGTGAGTTTCT | Sequencing for OsRR22 knockout target | |
| Cas9-F | CCTTCTGTGTGGTCTGGTTC | 载体检测 | |
| Cas9-R | AGACAATCACCCCCTGGAAC | Vector assay | |
| Hyg-F | CTATTTCTTTGCCCTCGGAC | 转基因检测 | |
| Hyg-R | ATGCCTGAACTCACCGCGAC | Transgenic detection | |
| qPCR SOS1-F | ATTGCAGCTGAGCATGTACG | 实时荧光定量PCR | |
| qPCR SOS1-R | AGAGCTTGCTTTCGTGTGAC | Quantitative real-time PCR | |
| qPCR OsMYB2-F | GGGCTGAAACGCACAGGCAAGA | 实时荧光定量PCR | |
| qPCR OsMYB2-R | CTGCTTGGCGTGCTTCTGC | Quantitative real-time PCR | |
| qPCR OsNHX1-F | GTGACAGACCTGGCAAATCC | 实时荧光定量PCR | |
| qPCR OsNHX1-R | TCGACACAGCTCCTCTCATC | Quantitative real-time PCR | |
| qPCR OsHKT1;1-F | TGCCAGAAGTTGTTGAAGCC | 实时荧光定量PCR | |
| qPCR OsHKT1;1-R | CCCAGGAACATCACCAGGAT | Quantitative real-time PCR | |
| qPCR OsP5CS1-F | GTCAGAGTGGACTGATGGCT | 实时荧光定量PCR | |
| qPCR OsP5CS1-R | GCCTTTCTAGTGCTGATGGC | Quantitative real-time PCR | |
| UBIQUITIN-F | GCTCCGTGGCGGTATCAT | 实时荧光定量PCR | |
| UBIQUITIN-R | CGGCAGTTGACAGCCCTAG | Quantitative real-time PCR | |
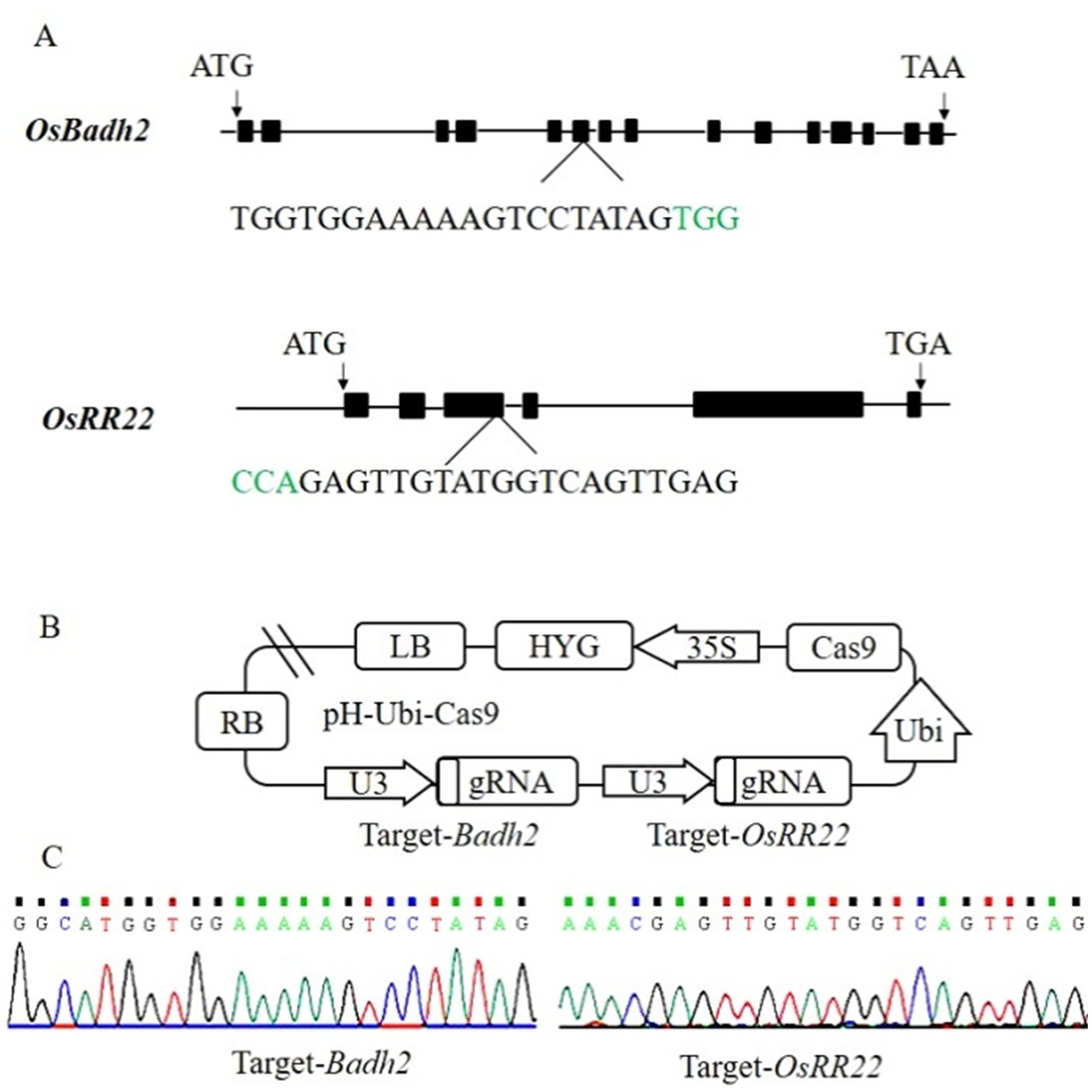
Fig. 1. Badh2 and OsRR22 target sites and CRISPR/Cas9 vector construction. A, Badh2 and OsRR22 structure and target site. The PAM motif is shown in green. B, Construction of CRISPR/Cas9 vector for Badh2 and OsRR22 gene target; C, Target site sequencing results in the expression vector.

Fig. 2. Identification of Cas9 markers in some T1 plants. M, DNA Marker; “+”, Positive control; “−”, Negative control; Lanes 1 to 9, Some plants of T1. The size of the Cas9 marker target fragment is 906 bp.

Fig. 3. Mutation types at the Badh2 and OsRR22 loci of the homozygous lines in T1. Red arrows indicate mutation positions. “+” means insertion; “-” means deletion. Black arrows indicate amino acid positions.

Fig. 4. Salt tolerance identification of T2 homozygous lines 21-30, 21-31 at the seedling stage. Values are shown as mean ± SD. *, ** Significantly different at 0.05 and 0.01 levels by t-test, respectively.
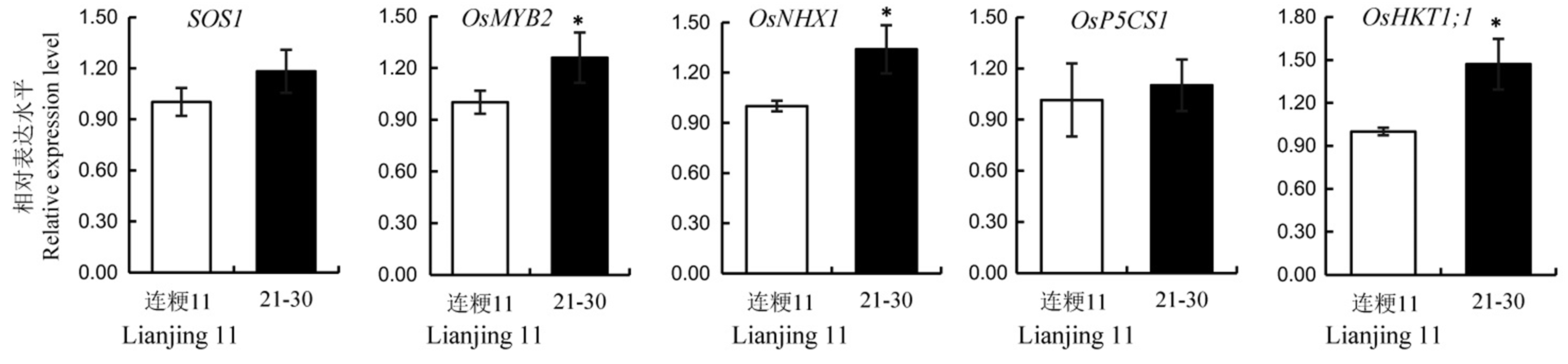
Fig. 5. Relative expression levels of salt resistance related genes in the wild-type and its T2 line 21-30. Values are shown as mean ± SD.*Significantly different at 0.05 level by t-test.
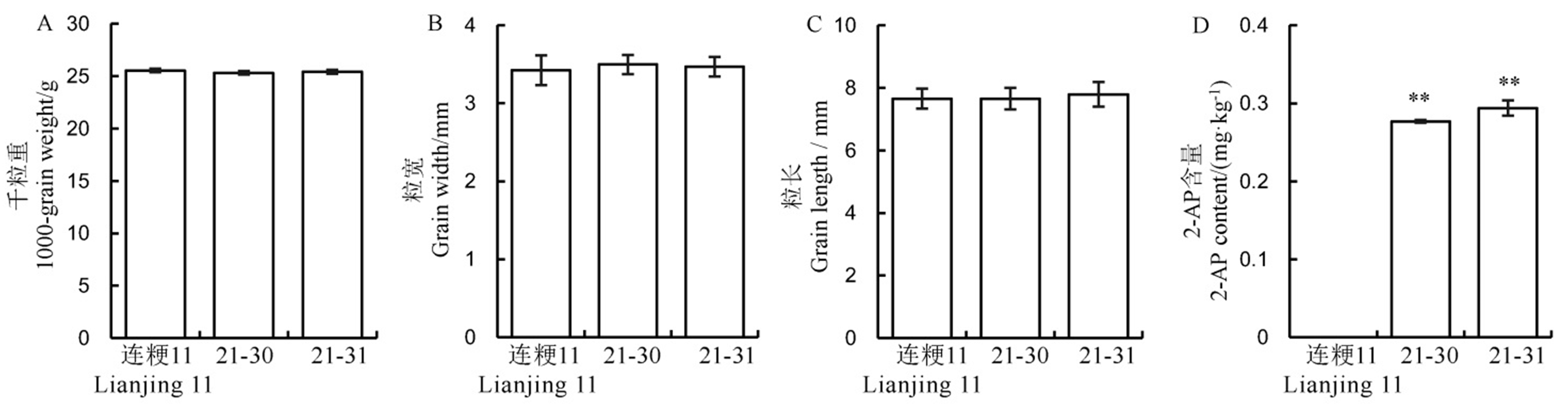
Fig. 6. Performance of rice yield and fragrance related traits in the wild-type and its positive transgenic lines. Values are shown as mean ± SD.** Significantly different at 0.01 level by t-test.
| [1] | El Mahi H, Perez-Hormaeche J, de Luca A, Villalta I, Espartero J, Gamez-Arjona F, Fernandez J L, Bundo M, Mendoza I, Mieulet D. A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice[J]. Plant Physiology, 2019, 180(2): 1046-1065. |
| [2] | Yang A, Dai X Y, Zhang W H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice[J]. Journal of Experimental Botany, 2012, 63(7): 2541-2556. |
| [3] | Wang R, Jing W, Xiao L G, Jin Y K, Shen L K, Zhang W H. The rice high-affinity potassium pransporter1;1 is involved in salt tolerance and regulated by an MYB-type transcription factor[J]. Plant Physiology, 2015, 168(3): 1076-1090. |
| [4] | Liu S P, Zheng L Q, Xue Y H, Zhang Q, Wang L, Shuo H X. Overexpression of OsVP1 and OsNHX1 increases tolerance to drought and salinity in rice[J]. Journal of Plant Biology, 2010, 53: 444-452. |
| [5] | 张霞, 唐维, 刘嘉, 刘永胜. 过量表达水稻 OsP5CS1 和OsP5CS2 基因提高烟草脯氨酸的生物合成及其非生物胁迫抗性[J]. 应用与环境生物学报, 2014, 20(4): 717-722. |
| Zhang X, Tang W, Liu J, Liu Y S. Co-expression of rice OsP5CS1 and OsP5CS2genes in transgenic tobacco resulted in elevated proline biosynthesis and enhanced abiotic stress tolerance[J]. Chinese Journal of Applied & Environmental Biology, 2014, 20(4): 717-722. (in Chinese with English abstract) | |
| [6] | Li H W, Wang X P. Overexpression of the trehalose- 6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice[J]. Planta, 2011, 234(5): 1007-1018. |
| [7] | Lan T, Zheng Y L, Su Z L, Yu S B, Song H B, Zheng X Y, Lin G G, Wu W R. OsSPL10, a SBP-box gene, plays a dual role in salt tolerance and trichome formation in rice (Oryza sativa L.)[J]. G3: Genes, Genomes, Genetics, 2019, 9(12): 4107-4114. |
| [8] | Yang C, Ma B, He S J, Xiong Q, Duan K X, Yin C C, Chen H, Lu X, Chen S Y, Zhang J S. MAOHUZI6/ETHYLENE INSENSITIVE3-LIKE1 and ETHYLENE INSENSITIVE3-LIKE2 regulate ethylene response of roots and coleoptiles and negatively affect salt tolerance in rice[J]. Plant Physiology, 2015, 169(1): 148-165. |
| [9] | Koh S, Lee S C, Kim M K, Koh J H, Lee S, An G, Choe S, Kim S R. T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses[J]. Plant Molecular Biology, 2007, 65(4): 453-466. |
| [10] | Takagi H, Tamiru M, Abe A, Yoshida K, Uemura A, Yaegashi H, Obara T, Oikawa K, Utsushi H, Kanzaki E, Mitsuoka C, Natsume S, Kosugi S, Kanzaki H, Matsumura H, Urasaki N, Kamoun S, Terauchi R. MutMap accelerates breeding of a salt-tolerant rice cultivar[J]. Nature Biotechnology, 2015, 33(5): 445-449. |
| [11] | Zhang A N, Liu Y, Wang F M, Li T F, Chen Z H, Kong D Y, Bi J G, Zhang F Y, Luo X X, Wang J H, Tang J J, Yu X Q, Liu G L, Luo L J. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene[J]. Molecular Breeding, 2019, 39: 47. |
| [12] | 李晶岚, 陈鑫欣, 石翠翠, 刘方惠, 孙静, 葛荣朝. OsRPK1基因过表达和RNA干涉对水稻苗期耐盐性的影响[J]. 作物学报, 2020, 46(8): 1217-1224. |
| Li J L, Chen X X, Shi C C, Liu F H, Sun J, Ge R C. Effects of OsRPK1 gene overexpression and RNAi on the salt-tolerance at seedling stage in rice[J]. Acta Agronomica Sinica, 2020, 46(8): 1217-1224. (in Chinese with English abstract) | |
| [13] | 魏晓东, 张亚东, 赵凌, 路凯, 宋雪梅, 王才林. 稻米香味物质2-乙酰-1-吡咯啉的形成及其影响因素[J]. 中国水稻科学, 2022, 36 (2): 131-138. |
| Wei X D, Zhang Y D, Zhao L, Lu K, Song X M, Wang C L. Research progress in biosynthesis and influencing factors of 2-acetyl-1-pyrroline in fragrant rice[J]. Chinese Journal of Rice Science, 2022, 36(2): 131-138. (in Chinese with English abstract) | |
| [14] | Jiang M, Liu Y H, Li R Q, Li S, Tan Y Y, Huang J Z, Shu Q Y. An inositol 1, 3, 4, 5, 6-pentakisphosphate 2-kinase 1 mutant with a 33-nt deletion showed enhanced tolerance to salt and drought stress in rice[J]. Plants, 2021, 10(1): 23. |
| [15] | Bradbury L M, Fitzgerald T L, Henry R J, Jin Q S, Waters D L. The gene for fragrance in rice[J]. Plant Biotechnology Journal, 2005, 3(3): 363-370. |
| [16] | Chen S H, Yang Y, Shi W W, Ji Q, He F, Zhang Z D, Heng Z K, Liu X N, Xu M L. Badh2, encoding betaine ldehyde dehydrogenase, inhibits the biosynthesis of -acetyl-1-pyrroline, a major component in rice fragrance[J]. Plant Cell, 2008, 20(7): 1850-1861. |
| [17] | Bradbury L M, Gillies S A, Brushett D J, Waters D L, Henry R J. Inactivation of an amino aldehyde dehydrogenase is responsible for fragrance in rice[J]. Plant Molecular Biology, 2008, 68(4-5): 439-449. |
| [18] | Shan Q W, Zhang Y, Chen K L, Zhang K, Gao C X. Creation of fragrant rice by targeted knockout of the OsBadh2 gene using TALEN technology[J]. Plant Biotechnology Journal, 2015, 13(6): 791-800. |
| [19] | Hui S Z, Li H J, Mawia A M, Zhou L, Cai J Y, Ahmad S, Lai C K, Wang J X, Jiao G A, Xie L H, Shao G N, Sheng Z H, Tang S Q, Wang J L, Wei X J, Hu S K, Hu P S. Production of aromatic three-line hybrid rice using novel alleles of BADH2[J]. Plant Biotechnology Journal, 2022, 20(1): 59-74. |
| [20] | Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu L. Targeted mutagenesis in rice using CRISPR-Cas system[J]. Cell Research, 2013, 23(10): 1233-1236. |
| [21] | Yoshida S, Forno D A, Cook J H, Gomez K A. Laboratory Manual for Physiological Studies of Rice[M]. International Rice Research Institute, Philippines, 1976: 61-66. |
| [22] | Liu Y, Wang B X, Li J, Song Z Q, Lu B G, Chi M, Yang B, Liu J B, Lam Y W, Li J X, Xu D Y. Salt-response analysis in two rice cultivars at seedling stage[J]. Acta Physiologiae Plantarum, 2017, 39: 215. |
| [23] | Wang B X, Xu B, Liu Y, Li J F, Sun Z G, Chi M, Xing Y G, Yang B, Li J, Liu J B, Lu B G, Xu D Y, Bello B K. A novel mechanism of the signaling cascade associated with the SAPK10-bZIP20-NHX1 synergistic interaction to enhance tolerance of plant to abiotic stress in rice[J]. Plant Science, 2022, 323: 111393. |
| [24] | Xu Y, Lin Q P, Li X F, Wang F J, Chen Z H, Wang J, Li W Q, Fan F J, Tao Y J, Jiang Y J, Wei X D, Zhang R, Yang J, Gao C X. Fine-tuning the amylose content of rice by precise base editing of the Wx gene[J]. Plant Biotechnology Journal, 2021, 19(1): 11-13. |
| [25] | Liu X X, Liu H L, Zhang Y Y, He M L Li R T, Meng W, Wang Z Y, Li X F, Bu Q Y. Fine-tuning flowering time via genome editing of upstream open reading frames of heading date 2 in rice[J]. Rice, 2021, 14(1): 59. |
| [26] | 徐善斌, 郑洪亮, 刘利锋, 卜庆云, 李秀峰, 邹德堂. 利用CRISPR/Cas9 技术高效创制长粒香型水稻[J]. 中国水稻科学, 2020, 34(5): 406-412. |
| Xu S B, Zheng H L, Liu L F, Bu Q Y, Li X F, Zou D T. Improvement of grain shape and fragrance by using CRISPR/Cas9 system[J]. Chinese Journal of Rice Science, 2020, 34(5): 406-412. (in Chinese with English abstract) |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||