
Chinese Journal OF Rice Science ›› 2023, Vol. 37 ›› Issue (2): 133-141.DOI: 10.16819/j.1001-7216.2023.220607
• Research Papers • Previous Articles Next Articles
WEI Minyi1, MA Zengfeng1, HUANG Dahui1, QIN Yuanyuan2, LIU Chi1, LU Yingping1,3, LUO Tongping1, LI Zhenjing1, ZHANG Yuexiong1,*( ), QIN Gang1,*(
), QIN Gang1,*( )
)
Received:2022-06-14
Revised:2022-08-05
Online:2023-03-10
Published:2023-03-10
Contact:
ZHANG Yuexiong, QIN Gang
韦敏益1, 马增凤1, 黄大辉1, 秦媛媛2, 刘驰1, 卢颖萍1,3, 罗同平1, 李振经1, 张月雄1,*( ), 秦钢1,*(
), 秦钢1,*( )
)
通讯作者:
张月雄,秦钢
基金资助:WEI Minyi, MA Zengfeng, HUANG Dahui, QIN Yuanyuan, LIU Chi, LU Yingping, LUO Tongping, LI Zhenjing, ZHANG Yuexiong, QIN Gang. QTL-Seq Analysis for Identification of Resistance Locus to Bacterial Leaf Streak in Rice[J]. Chinese Journal OF Rice Science, 2023, 37(2): 133-141.
韦敏益, 马增凤, 黄大辉, 秦媛媛, 刘驰, 卢颖萍, 罗同平, 李振经, 张月雄, 秦钢. 基于QTL-Seq的水稻抗细菌性条斑病QTL定位[J]. 中国水稻科学, 2023, 37(2): 133-141.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2023.220607
| 标记 | 物理位置 | 正向引物 | 反向引物 |
|---|---|---|---|
| Marker | Position / bp | Forward primer(5’-3’) | Reverse primer(5’-3’) |
| C4M1 | 557 006 | TATACACGCAACTCCCCCTC | TGCCAATACCACAGTCCTGA |
| C4M2 | 1 332 530 | TTTCTCCAAACCGAACAAGC | CAGTGCAGCAGGGAAGTACA |
| C4M3 | 2 519 322 | CTCCCACCTGACAAGTTCGT | CAACCAGAGAAAAGCAAGCA |
| C4M5 | 4 314 270 | CGAAAGGTCAAAACGTTGGT | GTGCATCCGCCTTAATTTGT |
| C4M6 | 5 006 279 | CGTCATGGTCATCGTACGTC | CACGTAACGCAACGGATATG |
| C4M8 | 5 534 085 | AATCACCGAGAGCATTTTGG | GTGCAACTTTTACGTGGCCT |
| C4M9 | 6 070 782 | AGAGACTGTCGCTCCCAAAA | GGGAAAGCTCTGAAATGATCC |
| C4M10 | 6 888 374 | GAGAGCCCGTAAACATTCCA | ACCATCAACCGGATTATTCA |
| C4M13 | 11 782 373 | CCCTGGAAAGAACAAACCAA | ATGACCGAGCCTGATATTGC |
| C4M14 | 13 911 219 | AGCATGTTAAATCATCATCCCA | TGGCTTAACGAAAGAAGAGAGAA |
| C4MJ1 | 5 269 417 | GGTCTCTCTCTCATGCGTCC | ACCAGACCAGGCAGCTAAGA |
| C4MJ5 | 5 364 530 | TTCCCCTAAACCATTTTTCTCTC | AGGAGGAAGGGAGGTGCTAC |
| C4MJ2 | 5 691 595 | CGTCAAGGAACACAGCGATA | TGGGACGGATGGATTAGTTT |
| C4MJ3 | 5 790 460 | GCCTGAATCACGAAGCTCAT | GAGAAGGTTTGCTCCTGCAC |
Table 1. InDel markers for gene mapping in this study.
| 标记 | 物理位置 | 正向引物 | 反向引物 |
|---|---|---|---|
| Marker | Position / bp | Forward primer(5’-3’) | Reverse primer(5’-3’) |
| C4M1 | 557 006 | TATACACGCAACTCCCCCTC | TGCCAATACCACAGTCCTGA |
| C4M2 | 1 332 530 | TTTCTCCAAACCGAACAAGC | CAGTGCAGCAGGGAAGTACA |
| C4M3 | 2 519 322 | CTCCCACCTGACAAGTTCGT | CAACCAGAGAAAAGCAAGCA |
| C4M5 | 4 314 270 | CGAAAGGTCAAAACGTTGGT | GTGCATCCGCCTTAATTTGT |
| C4M6 | 5 006 279 | CGTCATGGTCATCGTACGTC | CACGTAACGCAACGGATATG |
| C4M8 | 5 534 085 | AATCACCGAGAGCATTTTGG | GTGCAACTTTTACGTGGCCT |
| C4M9 | 6 070 782 | AGAGACTGTCGCTCCCAAAA | GGGAAAGCTCTGAAATGATCC |
| C4M10 | 6 888 374 | GAGAGCCCGTAAACATTCCA | ACCATCAACCGGATTATTCA |
| C4M13 | 11 782 373 | CCCTGGAAAGAACAAACCAA | ATGACCGAGCCTGATATTGC |
| C4M14 | 13 911 219 | AGCATGTTAAATCATCATCCCA | TGGCTTAACGAAAGAAGAGAGAA |
| C4MJ1 | 5 269 417 | GGTCTCTCTCTCATGCGTCC | ACCAGACCAGGCAGCTAAGA |
| C4MJ5 | 5 364 530 | TTCCCCTAAACCATTTTTCTCTC | AGGAGGAAGGGAGGTGCTAC |
| C4MJ2 | 5 691 595 | CGTCAAGGAACACAGCGATA | TGGGACGGATGGATTAGTTT |
| C4MJ3 | 5 790 460 | GCCTGAATCACGAAGCTCAT | GAGAAGGTTTGCTCCTGCAC |
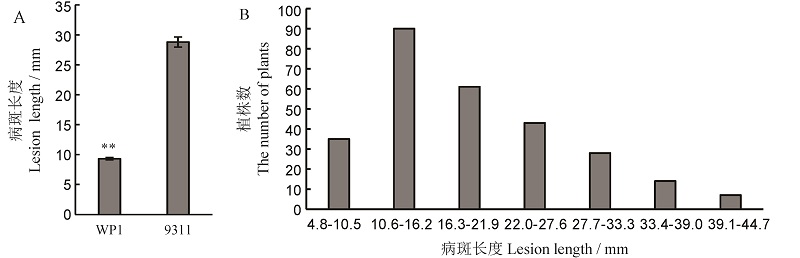
Fig. 2. Resistance of parents to bacterial leaf streak(A) and distribution frequency of lesion length in F8:9 population(B). ** Difference between the parents was significant at 0.01 level.
| 关联分析方法 Association analysis method | 染色体 Chromosome | 起始Start | 终止End | 片段大小 Size/Mb | 基因数量 Gene number |
|---|---|---|---|---|---|
| 基于SNP的ED方法关联结果 Results of ED method based on SNP | 10 | 16,070,000 | 21,090,000 | 5.02 | 925 |
| 4 | 6,170,000 | 6,840,000 | 0.67 | 99 | |
| 8 | 20,840,000 | 24,160,000 | 3.32 | 547 | |
| 基于SNP的ΔSNP-index方法关联结果 Results of ΔSNP-index method based on SNP | 10 | 17,060,000 | 20,860,000 | 3.80 | 697 |
| 基于InDel的ED方法关联结果 Results of ED method based on InDel | 10 | 16,180,000 | 21,160,000 | 4.98 | 919 |
| 4 | 5,640,000 | 7,520,000 | 1.88 | 244 | |
| 8 | 20,650,000 | 24,170,000 | 3.52 | 575 | |
| 基于InDel的ΔSNP-index方法关联结果 Results of SNP-index method based on InDel | 10 | 17,280,000 | 21,000,000 | 3.72 | 693 |
Table 2. Results of QTL-seq.
| 关联分析方法 Association analysis method | 染色体 Chromosome | 起始Start | 终止End | 片段大小 Size/Mb | 基因数量 Gene number |
|---|---|---|---|---|---|
| 基于SNP的ED方法关联结果 Results of ED method based on SNP | 10 | 16,070,000 | 21,090,000 | 5.02 | 925 |
| 4 | 6,170,000 | 6,840,000 | 0.67 | 99 | |
| 8 | 20,840,000 | 24,160,000 | 3.32 | 547 | |
| 基于SNP的ΔSNP-index方法关联结果 Results of ΔSNP-index method based on SNP | 10 | 17,060,000 | 20,860,000 | 3.80 | 697 |
| 基于InDel的ED方法关联结果 Results of ED method based on InDel | 10 | 16,180,000 | 21,160,000 | 4.98 | 919 |
| 4 | 5,640,000 | 7,520,000 | 1.88 | 244 | |
| 8 | 20,650,000 | 24,170,000 | 3.52 | 575 | |
| 基于InDel的ΔSNP-index方法关联结果 Results of SNP-index method based on InDel | 10 | 17,280,000 | 21,000,000 | 3.72 | 693 |
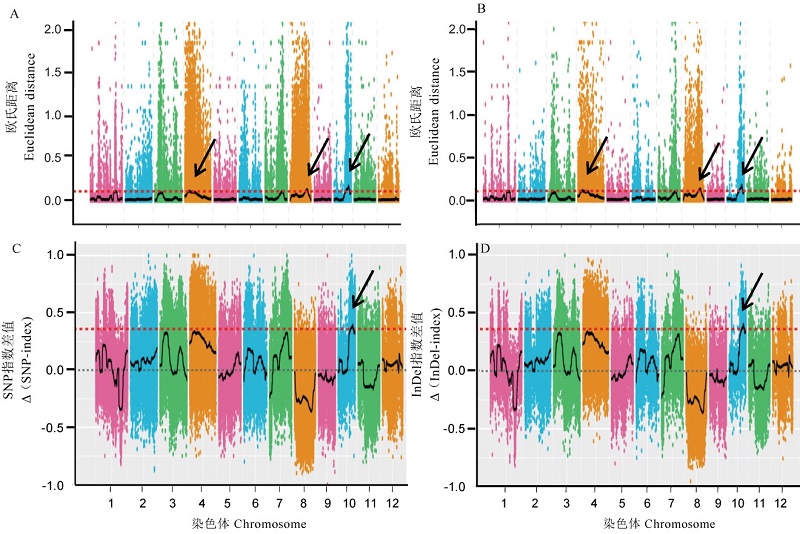
Fig. 3. ED and △(SNP-index) correlation analysis based on SNP and InDel of BSA-seq. A and B, ED association analysis figures based on SNP and InDel, respectively. C and D, △ (SNP-index) correlation analysis diagram based on SNP and InDel, respectively. The red dotted line is the 99% threshold line.
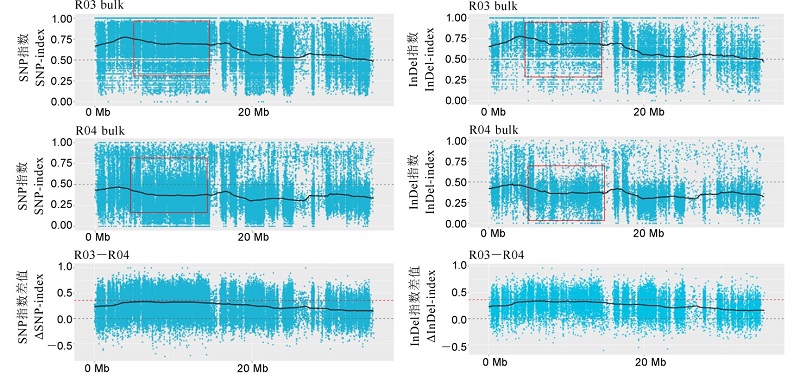
Fig. 4. SNP-index and InDel -index of the resistant pool and the susceptible pool on chromosome 4. A, B are the distribution of SNP-index values in resistant and susceptible mixed pools, respectively; C shows the distribution of Δ(SNP-index) value. The red dotted line is the 99% threshold line. R03, Resistant pool; R04, Susceptible pool.
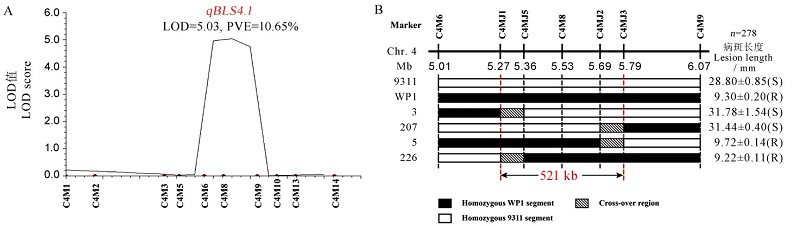
Fig. 5. Verification and fine mapping of the resistance locus qBLS4.1. A, Location and genetic effect of qBLS4.1; B, Graphical genotypes and resistance phenotypes of the recombinants.
| ORF编号 | 基因 | 突变类型 | 基因功能注释 |
|---|---|---|---|
| Accession | Gene | Mutation type | Putative function |
| 1 | LOC_Os04g09870 | SNP | 假设蛋白 Hypothetical protein |
| 2 | LOC_Os04g09880 | SNP | 表达蛋白 Expressed protein |
| 3 | LOC_Os04g09900 | SNP | 贝壳杉烯合酶,叶绿体前体 Ent-kaurene synthase, chloroplast precursor |
| 4 | LOC_Os04g09910 | SNP | 反转录转座子蛋白,Ty3-gypsy亚类 Retrotransposon protein, Ty3-gypsy subclass |
| 5 | LOC_Os04g09920 | InDel/SNP | 细胞色素P450 Cytochrome P450 |
| 6 | LOC_Os04g09950 | SNP | 假设蛋白 Hypothetical protein |
| 7 | LOC_Os04g09960 | InDel | 表达蛋白 Expressed protein |
| 8 | LOC_Os04g09990 | InDel | 假设蛋白 Hypothetical protein |
| 9 | LOC_Os04g10010 | InDel/SNP | 表达蛋白 Expressed protein |
| 10 | LOC_Os04g10070 | SNP | 反转录转座子蛋白,Ty3-gypsy亚类 Retrotransposon protein, Ty3-gypsy subclass |
| 11 | LOC_Os04g10100 | SNP | 反转录转座子蛋白,Ty3-gypsy亚类 Retrotransposon protein, Ty3-gypsy subclass |
| 12 | LOC_Os04g10110 | SNP | 反转录转座子蛋白,Ty3-gypsy亚类 Retrotransposon protein, Ty3-gypsy subclass |
| 13 | LOC_Os04g10120 | SNP | 反转录转座子蛋白,Ty3-gypsy亚类 Retrotransposon protein, Ty3-gypsy subclass |
| 14 | LOC_Os04g10140 | SNP | 假设蛋白 Hypothetical protein |
| 15 | LOC_Os04g10160 | SNP | 细胞色素P450 Cytochrome P450 |
| 16 | LOC_Os04g10170 | SNP | 假设蛋白 Hypothetical protein |
| 17 | LOC_Os04g10180 | InDel/SNP | 表达蛋白 Expressed protein |
| 18 | LOC_Os04g10190 | InDel/SNP | 反转录转座子蛋白,Ty3-gypsy亚类 Retrotransposon protein, Ty3-gypsy subclass |
| 19 | LOC_Os04g10200 | SNP | 反转录转座子蛋白,Ty3-gypsy亚类 Retrotransposon protein, Ty3-gypsy subclass |
| 20 | LOC_Os04g10214 | SNP | 表达蛋白 Expressed protein |
| 21 | LOC_Os04g10219 | InDel | 表达蛋白 Expressed protein |
| 22 | LOC_Os04g10230 | SNP | 表达蛋白 Expressed protein |
| 23 | LOC_Os04g10240 | SNP | 表达蛋白 Expressed protein |
| 24 | LOC_Os04g10260 | SNP | 含碱性亮氨酸拉链结构域的蛋白 Basic region leucine zipper domain containing protein |
| 25 | LOC_Os04g10270 | InDel/SNP | 反转录转座子蛋白 Retrotransposon protein, unclassified |
| 26 | LOC_Os04g10290 | SNP | 表达蛋白 Expressed protein |
| 27 | LOC_Os04g10300 | SNP | 假设蛋白 Hypothetical protein |
| 28 | LOC_Os04g10320 | SNP | 表达蛋白 Expressed protein |
| 29 | LOC_Os04g10330 | SNP | 表达蛋白 Expressed protein |
| 30 | LOC_Os04g10390 | InDel/SNP | 反转录转座子蛋白 Retrotransposon protein, unclassified |
| 31 | LOC_Os04g10400 | SNP | 电子转运黄素蛋白亚单位β Electron transfer flavoprotein subunit beta |
| 32 | LOC_Os04g10410 | SNP | 酰胺酶 Amidase |
| 33 | LOC_Os04g10420 | InDel/SNP | CW7 |
| 34 | LOC_Os04g10450 | SNP | 假设蛋白 Hypothetical protein |
| 35 | LOC_Os04g10500 | SNP | 表达蛋白 Expressed protein |
| 36 | LOC_Os04g10550 | InDel/SNP | 表达蛋白 Expressed protein |
| 37 | LOC_Os04g10569 | SNP | 谷氨酰-tRNA酰胺转移酶 Glutamyl-tRNA amidotransferase |
| 38 | LOC_Os04g10590 | SNP | 反转录转座子蛋白 Retrotransposon protein, unclassified |
| 39 | LOC_Os04g10620 | SNP | 反转录转座子蛋白,Ty3-gypsy亚类 Retrotransposon protein, Ty3-gypsy subclass |
| 40 | LOC_Os04g10630 | InDel/SNP | 表达蛋白 Expressed protein |
| 41 | LOC_Os04g10650 | SNP | CDT1A-DNA复制起始蛋白 CDT1A-DNA replication initiation protein |
Table 3. Candidate gene annotation in the location interval.
| ORF编号 | 基因 | 突变类型 | 基因功能注释 |
|---|---|---|---|
| Accession | Gene | Mutation type | Putative function |
| 1 | LOC_Os04g09870 | SNP | 假设蛋白 Hypothetical protein |
| 2 | LOC_Os04g09880 | SNP | 表达蛋白 Expressed protein |
| 3 | LOC_Os04g09900 | SNP | 贝壳杉烯合酶,叶绿体前体 Ent-kaurene synthase, chloroplast precursor |
| 4 | LOC_Os04g09910 | SNP | 反转录转座子蛋白,Ty3-gypsy亚类 Retrotransposon protein, Ty3-gypsy subclass |
| 5 | LOC_Os04g09920 | InDel/SNP | 细胞色素P450 Cytochrome P450 |
| 6 | LOC_Os04g09950 | SNP | 假设蛋白 Hypothetical protein |
| 7 | LOC_Os04g09960 | InDel | 表达蛋白 Expressed protein |
| 8 | LOC_Os04g09990 | InDel | 假设蛋白 Hypothetical protein |
| 9 | LOC_Os04g10010 | InDel/SNP | 表达蛋白 Expressed protein |
| 10 | LOC_Os04g10070 | SNP | 反转录转座子蛋白,Ty3-gypsy亚类 Retrotransposon protein, Ty3-gypsy subclass |
| 11 | LOC_Os04g10100 | SNP | 反转录转座子蛋白,Ty3-gypsy亚类 Retrotransposon protein, Ty3-gypsy subclass |
| 12 | LOC_Os04g10110 | SNP | 反转录转座子蛋白,Ty3-gypsy亚类 Retrotransposon protein, Ty3-gypsy subclass |
| 13 | LOC_Os04g10120 | SNP | 反转录转座子蛋白,Ty3-gypsy亚类 Retrotransposon protein, Ty3-gypsy subclass |
| 14 | LOC_Os04g10140 | SNP | 假设蛋白 Hypothetical protein |
| 15 | LOC_Os04g10160 | SNP | 细胞色素P450 Cytochrome P450 |
| 16 | LOC_Os04g10170 | SNP | 假设蛋白 Hypothetical protein |
| 17 | LOC_Os04g10180 | InDel/SNP | 表达蛋白 Expressed protein |
| 18 | LOC_Os04g10190 | InDel/SNP | 反转录转座子蛋白,Ty3-gypsy亚类 Retrotransposon protein, Ty3-gypsy subclass |
| 19 | LOC_Os04g10200 | SNP | 反转录转座子蛋白,Ty3-gypsy亚类 Retrotransposon protein, Ty3-gypsy subclass |
| 20 | LOC_Os04g10214 | SNP | 表达蛋白 Expressed protein |
| 21 | LOC_Os04g10219 | InDel | 表达蛋白 Expressed protein |
| 22 | LOC_Os04g10230 | SNP | 表达蛋白 Expressed protein |
| 23 | LOC_Os04g10240 | SNP | 表达蛋白 Expressed protein |
| 24 | LOC_Os04g10260 | SNP | 含碱性亮氨酸拉链结构域的蛋白 Basic region leucine zipper domain containing protein |
| 25 | LOC_Os04g10270 | InDel/SNP | 反转录转座子蛋白 Retrotransposon protein, unclassified |
| 26 | LOC_Os04g10290 | SNP | 表达蛋白 Expressed protein |
| 27 | LOC_Os04g10300 | SNP | 假设蛋白 Hypothetical protein |
| 28 | LOC_Os04g10320 | SNP | 表达蛋白 Expressed protein |
| 29 | LOC_Os04g10330 | SNP | 表达蛋白 Expressed protein |
| 30 | LOC_Os04g10390 | InDel/SNP | 反转录转座子蛋白 Retrotransposon protein, unclassified |
| 31 | LOC_Os04g10400 | SNP | 电子转运黄素蛋白亚单位β Electron transfer flavoprotein subunit beta |
| 32 | LOC_Os04g10410 | SNP | 酰胺酶 Amidase |
| 33 | LOC_Os04g10420 | InDel/SNP | CW7 |
| 34 | LOC_Os04g10450 | SNP | 假设蛋白 Hypothetical protein |
| 35 | LOC_Os04g10500 | SNP | 表达蛋白 Expressed protein |
| 36 | LOC_Os04g10550 | InDel/SNP | 表达蛋白 Expressed protein |
| 37 | LOC_Os04g10569 | SNP | 谷氨酰-tRNA酰胺转移酶 Glutamyl-tRNA amidotransferase |
| 38 | LOC_Os04g10590 | SNP | 反转录转座子蛋白 Retrotransposon protein, unclassified |
| 39 | LOC_Os04g10620 | SNP | 反转录转座子蛋白,Ty3-gypsy亚类 Retrotransposon protein, Ty3-gypsy subclass |
| 40 | LOC_Os04g10630 | InDel/SNP | 表达蛋白 Expressed protein |
| 41 | LOC_Os04g10650 | SNP | CDT1A-DNA复制起始蛋白 CDT1A-DNA replication initiation protein |
| [1] | Yang W, Ju Y H, Zuo L P, Shang L Y, Li X R, Li X M, Feng S Z, Ding X H, Chu Z H. OsHsfB4d binds the promoter and 11/11 regulates the expression of OsHsp18.0-CI to resistant against Xanthomonas oryzae[J]. Rice, 2020, 13(1): 28. |
| [2] | Liu W, Liu J, Triplett L, Leach J E, Wang G L. Novel insights into rice innate immunity against bacterial and fungal pathogens[J]. Annual Review of Phytopathology, 2013, 52: 213-241. |
| [3] | He W A, Huang D H, Li R B, Qiu Y F, Song J D, Yang H N, Zheng J X, Huang Y Y, Liu C, Zhang Y X, Ma Z F, Yang Y. Identification of a resistance gene bls1 to bacterial leaf streak in wild rice Oryza rufipogon Griff[J]. Journal of Integrative Agriculture, 2012, 11: 962-969. |
| [4] | Ma Z F, Qin G, Zhang Y X, Liu C, Wei M Y, Cen Z L, Yan Y, Luo T P, Li Z J, Liang H F, Huang D H, Deng G F. Bacterial Leaf Streak 1 encoding a mitogen-activated protein kinase confers the rice resistance to bacterial leaf streak[J]. The Plant Journal, 2021, 107(4): 1084-1101. |
| [5] | 吴为人, 唐定中, 李维明, 卢浩然, Worland A J. 水稻细菌性条斑病抗性基因定位[J]. 高技术通讯, 1998(7): 47-50. |
| Wu W R, Tang D Z, Li W M, Lu H R, Worland A J. Mapping of genes underlying resistance to bacterial leaf streak in rice[J]. Chinese High Technology Letters, 1998(7): 47-50. (in Chinese with English abstract) | |
| [6] | Tang D Z, Wu W R, Li W M, Lu H R, Worland A J. Mapping of QTLs conferring resistance to bacterial leaf streak in rice[J]. Theoretical and Applied Genetics, 2000, 101: 286-291. |
| [7] | 陈志伟, 景艳军, 李小辉, 周元昌, 刁志娟, 李生平, 吴为人. 水稻细菌性条斑病抗性QTL qBlsr5a的验证和更精确定位[J]. 福建农林大学学报:自然科学版, 2006, 35(6): 619-622. |
| Chen Z W, Jing Y J, Li X H, Zhou Y C, Diao Z J, Li S P, Wu W R. Verification and more precise mapping of a QTL qBIsr5a underlying resistance to bacterial leaf streak in rice[J]. Journal of Fujian Agriculture and Forestry University: Natural Science, 2006, 35(6): 619-622. (in Chinese with English abstract) | |
| [8] | Han Q D, Chen Z W, Deng Y L, Lan T, Guan H Z, Guan Y L, Zhou Y C, Lin M C, Wu W R. Fine mapping of qBlsr5a, a QTL controlling resistance to bacterial leaf streak in rice[J]. Acta Agronomica Sinica, 2008, 34(4): 587-590. |
| [9] | Xie X F, Chen Z W, Cao J L, Guan H Z, Lin D G, Li C L, Lan T, Duan Y L, Mao D M, Wu W R. Toward the positional cloning of qBlsr5a, a QTL underlying resistance to bacterial leaf streak, using overlapping sub-CSSLs in rice[J]. PLoS ONE, 2014, 9(4): e95751. |
| [10] | Triplett L R, Cohen S P, Heffelfinger C, Schmidt C L, Huerta A I, Tekete C, Verdier V, Bogdanove A J, Leach J E. A resistance locus in the American heirloom rice variety carolina gold select is triggered by TAL effectors with diverse predicted targets and is effective against African strains of Xanthomonas oryzae pv. oryzicola[J]. The Plant Journal, 2016, 87(5): 472-483. |
| [11] | 施力军, 罗登杰, 赵严, 岑贞陆, 刘芳, 李容柏. 普通野生稻抗细菌性条斑病基因的遗传分析与定位[J]. 华南农业大学学报, 2019, 40(2): 1-5. |
| Shi L J, Luo D J, Zhao Y, Cen Z L, Liu F, Li R B. Genetic analysis and mapping of bacterial leaf streak resistance genes in Oryzae rufipogon Griff[J]. Journal of South China Agricultural University, 2019, 40(2): 1-5. (in Chinese with English abstract) | |
| [12] | 罗登杰, 万瑶, 覃雪梅, 施力军, 张慧, 李容柏, 刘芳. 水稻细菌性条斑病抗性基因bls2 SSR分子标记开发[J]. 南方农业学报, 2021, 52(5): 1167-1173. |
| Luo D J, Wan Y, Qin X M, Shi L J, Zhang H, Li R B, Liu F. Development of SSR molecular markers for bacterial leaf streak resistance gene bls2 in rice(Oryza sativa L.)[J]. Journal of Southern Agriculture, 2021, 52(5): 1167-1173. (in Chinese with English abstract) | |
| [13] | Wu T, Peng C, Li B B, Wu W, Kong L G, Li F C, Chu Z H, Liu F, Ding X H. OsPGIP1-mediated resistance to bacterial leaf streak in rice is beyond responsive to the polygalacturonase of Xanthomonas oryzae pv. oryzicola[J]. Rice, 2019, 12: 90. |
| [14] | Shen X L, Yuan B, Liu H B, Li X H, Wang S P. Opposite functions of a rice mitogen-activated protein kinase during the process of resistance against Xanthomonas oryzae[J]. The Plant Journal, 2010, 64(1): 86-99. |
| [15] | Feng C S, Zhang X, Wu T, Yuan B, Ding X H, Yao F Y, Chu Z H. The polygalacturonase-inhibiting protein 4 (OsPGIP4), a potential component of the qBlsr5a locus, confers resistance to bacterial leaf streak in rice[J]. Planta, 2016, 243: 1297-1308. |
| [16] | Ma H G, Chen J, Zhang Z Z, Ma L, Yang Z Y, Zhang Q L, Li X H, Xiao J H, Wang S P. MAPK kinase 10.2 promotes disease resistance and drought tolerance by activating different MAPKs in rice[J]. The Plant Journal, 2017, 92: 557-570. |
| [17] | Ju Y H, Tian H J, Zhang R H, Zuo L P, Jin G X, Xu Q, Ding X H, Li X K, Chu Z H. Overexpression of OsHSP18.0-CI enhances resistance to bacterial leaf streak in rice[J]. Rice, 2017, 10: 12. |
| [18] | Yang W, Zhang B G, Qi G H, Shang L Y, Chu Z H. Identification of the phytosulfokine receptor 1 (OsPSKR1) confers resistance to bacterial leaf streak in rice[J]. Planta, 2019, 250: 1603-1612. |
| [19] | Jiang N, Yan J, Liang Y, Shi Y L, He Z Z, Wu Y T, Zeng Q, Liu X L, Peng J H. Resistance genes and their interactions with bacterial blight/leaf streak pathogens (Xanthomonas oryzae) in rice (Oryza sativa L.): An updated review[J]. Rice, 2020, 13: 3. |
| [20] | Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform[J]. Bioinformatics, 2009, 14(1): 1754-1760. |
| [21] | Mckenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristol M A. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data[J]. Genome Research, 2010, 20(9): 1297-1303. |
| [22] | Mansfeld B N, Grumet R. QTLseqr: An R package for bulk segregant analysis with next-generation sequencing[J]. Plant Genome, 2018, 11(2): 1-5. |
| [23] | Paterson A H, DeVerna J W, Lanini B, Tanksley S D. Fine mapping of quantitative trait loci using selected overlapping recombinant chromosomes, in an interspecies cross of tomato[J]. Genetics, 1990, 124: 735-742. |
| [24] | 彭小群, 王梦龙. 水稻白叶枯病抗性基因研究进展[J]. 植物生理学报, 2022, 58(3): 472-482. |
| Peng X Q, Wang M L. Research advances on resistance genes to bacterial blight disease in rice[J]. Plant Physiology Journal, 2022, 58(3): 472-482. (in Chinese with English abstract) | |
| [25] | 李仲惺, 楼珏, 卢华金. 水稻细菌性条斑病发病程度与发病因子关系探讨[J]. 中国稻米, 2016, 22(4): 62-64. |
| Li Z X, Lou J, Lu H. Relationship between incidence and disease factors of bacterial leaf streak in rice[J]. China Rice, 2016, 22(4): 62-64. (in Chinese with English abstract) | |
| [26] | Xie X F, Zheng Y, Lu L B, Yuan J Z, Hu J, Bu S H, Lin Y Y, Liu Y S, Guan H Z, Wu W R. Genome-wide association study of QTLs conferring resistance to bacterial leaf streak in rice[J]. Plants (Basel), 2021, 10(10): 2039. |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||