
Chinese Journal OF Rice Science ›› 2023, Vol. 37 ›› Issue (2): 113-124.DOI: 10.16819/j.1001-7216.2023.220314
• Research Papers • Next Articles
LIAN Yuanxun1,2,#, WEI Ziyun1,2,#, ZHANG Qiang2, LI Qing2, REN Deyong2, HU Jiang2, ZHU Li2, GAO Zhenyu2, ZHANG Guangheng2, GUO Longbiao2, ZENG Dali2, QIAN Qian2,*( ), SHEN Lan2,*(
), SHEN Lan2,*( )
)
Received:2022-03-28
Revised:2022-05-09
Online:2023-03-10
Published:2023-03-10
Contact:
QIAN Qian, SHEN Lan
About author:First author contact:#These authors contributed equally to this work
廉院训1,2,#, 韦子芸1,2,#, 张强2, 李清2, 任德勇2, 胡江2, 朱丽2, 高振宇2, 张光恒2, 郭龙彪2, 曾大力2, 钱前2,*( ), 沈兰2,*(
), 沈兰2,*( )
)
通讯作者:
钱前,沈兰
作者简介:第一联系人:#共同第一作者
基金资助:LIAN Yuanxun, WEI Ziyun, ZHANG Qiang, LI Qing, REN Deyong, HU Jiang, ZHU Li, GAO Zhenyu, ZHANG Guangheng, GUO Longbiao, ZENG Dali, QIAN Qian, SHEN Lan. Identification and Gene Mapping of a Zebra Leaf Mutant zl7 in Rice[J]. Chinese Journal OF Rice Science, 2023, 37(2): 113-124.
廉院训, 韦子芸, 张强, 李清, 任德勇, 胡江, 朱丽, 高振宇, 张光恒, 郭龙彪, 曾大力, 钱前, 沈兰. 水稻斑马叶突变体zl7的鉴定与基因的精细定位[J]. 中国水稻科学, 2023, 37(2): 113-124.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2023.220314
| 标记 Marker | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|
| M1 | GTCCATGCATCCATCTCTAG | ACGGAAGGAATACGTCTGTA |
| M2 | TGTGGACAACCTCAACTGAAAGC | CATAATCACCAACATCGGAGAAGC |
| M3 | TGTTGAGCTAGAAGAGAGGGG | TGAACACAAAAGGATGCGCT |
| M4 | CCAAGTCTTAAGCTACCCCT | CGCAGGGCTTAATAGAATAC |
| M5 | GATAGAGCGAGTGAGCAAAC | CCTACCAATTCAACTCCAAC |
| M6 | TGGATTGAGGATCAGGATAG | TCTGGAATTTTCCCTAATGA |
| M7 | GAAATCAGTCAGAAAGACCG | CCATCTTCTCACTGTGGAGT |
| M8 | GCCCTAATTGCTCCAGGTCT | AATTCTAGCAGTGTTCCATTGTG |
| M9 | TTCTTCCATGTAGCAAGCATT | CCTACTGCCTGCCAAATCTAT |
Table 1. Primers and sequences used in gene mapping.
| 标记 Marker | 正向引物序列 Forward primer sequence | 反向引物序列 Reverse primer sequence |
|---|---|---|
| M1 | GTCCATGCATCCATCTCTAG | ACGGAAGGAATACGTCTGTA |
| M2 | TGTGGACAACCTCAACTGAAAGC | CATAATCACCAACATCGGAGAAGC |
| M3 | TGTTGAGCTAGAAGAGAGGGG | TGAACACAAAAGGATGCGCT |
| M4 | CCAAGTCTTAAGCTACCCCT | CGCAGGGCTTAATAGAATAC |
| M5 | GATAGAGCGAGTGAGCAAAC | CCTACCAATTCAACTCCAAC |
| M6 | TGGATTGAGGATCAGGATAG | TCTGGAATTTTCCCTAATGA |
| M7 | GAAATCAGTCAGAAAGACCG | CCATCTTCTCACTGTGGAGT |
| M8 | GCCCTAATTGCTCCAGGTCT | AATTCTAGCAGTGTTCCATTGTG |
| M9 | TTCTTCCATGTAGCAAGCATT | CCTACTGCCTGCCAAATCTAT |
| 基因 Gene | 基因登录号 Gene ID | 定量正向引物序列 Forward primer sequence | 定量反向引物序列 Reverse primer sequence | 功能 Function |
|---|---|---|---|---|
| NYC1 | LOC_Os01g12710 | CATGCAACACCAACAAAAGG | GACCATTCCAGGAGAAGCAG | 叶绿素b还原酶基因 |
| NOL | LOC_Os03g45194 | CCACGAAAGGTATAGGATATG | TCAAGTCAGTCACCGCAGAT | 叶绿素b还原酶基因 |
| NYC3 | LOC_Os06g24730 | TCTATCTAGGTGCCAAAGGC | ATTCTGGCACCTGCTGTTTC | α/β折叠水解酶家族蛋白 |
| PAO | LOC_Os03g05310 | AAGCCTCCGATGTTACCGAA | CGAGGGTTTCCAGAATTTGA | 脱镁叶绿酸a加氧酶,叶绿体前体 |
| SGR | LOC_Os09g36200 | GCAATGTCGCCAAATGACG | GCTCACCACACTCATTCCCTAAAG | 镁离子去螯合酶 |
| RCCR1 | LOC_Os10g25020 | GGATCGACGATTGATTTCATG | GTCGAGGCGTTCAGAAAGAT | 红色叶绿素分解代谢还原酶 |
| RCCR2 | LOC_Os10g25040 | TGGCGAGGGACAGGAAGGT | GGATGTGGTGGCGAGAGAAAC | 红色叶绿素分解代谢还原酶 |
| LchP2 | LOC_Os09g17740 | GAAGAAGATCAAGAACGGCC | TTGCCGGGGACGAAGTTGGT | 捕光叶绿素a/b结合蛋白基因 |
| PsbA | LOC_Os12g19580 | AGAGACGCGAAAGTACAAGC | AAGTTGCGGTCAATAAGGTA | 光合反应中心蛋白 |
| RpoC2 | LOC_Os04g16830 | ATGCATCGCAGGTACACCAA | CCCTCGCGTAAATTGCTTTG | DNA定向RNA聚合酶亚单位β |
| Rps15 | LOC_Os12g10580 | AGATACGGAGACTTGCTTCA | GCTCCCTAATATCCAACTGACT | 核酮糖二磷酸羧化酶大链前体 |
| V1 | LOC_Os03g45400 | AGAATCAGCGCGAGAAGAGAACCT | TACACCAGCTTTGGAGGAGCTGAA | 质体蛋白 |
| V2 | LOC_Os03g20460 | AGCAGATCCGTGATTACATGGCGA | TGCCTCTTCACTCTCTGCAACCAA | 鸟苷酸激酶 |
| YGL8 | LOC_Os01g17170 | TGGATCTAACATGACACGCACCCA | ACTGTAACGGCATTCTTCTCCGGT | 镁原卟啉IX单酯环化酶的催化亚基 |
| CAO1 | LOC_Os10g41780 | TTGGCTCAGTTAATGAGGGCAGAATCC | GGATGCGCACGTTGAGCATCTTTGTGG | 叶绿素a加氧酶 |
| PORA | LOC_Os04g58200 | ATGGCTCTCCAAGTTCAG | TGGCTCACGCTAAGGAAC | NADPH:原叶绿素酸酯氧化还原酶A |
| PORB | LOC_Os10g35370 | CCGCAAGGAGGGAGCGGTG | CCCTCTTGGTGCTAAGGCCG | 原叶绿素酸酯氧化还原酶B |
| CHLH | LOC_Os03g20700 | GCACGGGAACTTGGCGTTTCATTA | ACATGTCCTGGAGCTGCTTCTCAT | 镁离子螯合酶H亚基 |
| CHLD | LOC_Os03g59640 | TAGCACAGCTGTCAGAGTGGGTTT | TTGCCAGCCACCTCAAGTATCTCA | 镁离子螯合酶D亚基 |
| CHLI | LOC_Os03g36540 | AGGGATGCTGAACTCAGGGTGAAA | AAGTAGGACTCACGGAACGCCTTT | 镁离子螯合酶I亚基 |
| DVR | LOC_Os03g22780 | AGCCCAGGTTCATCAAGGT | TGATCACCCTCTCGAAGAACT | 联乙烯还原酶基因 |
| OsCHLM | LOC_Os06g04150 | GCTTCATCTCCACGCAGTTCTACT | GCAATGACGAATCGAAGACGCACA | 镁原卟啉O-甲基转移酶 |
| YGL1 | LOC_Os05g28200 | CCAGCCACTGATGAAAGCAGCAAT | AGAGCGCTAATACACTCGCGAACA | 叶绿素合成酶 |
| OsHEMA1 | LOC_Os10g35840 | GATGCAATCACTGCTGGAAAGCGT | CCATCTTGCCAGCACCAATCAACA | 莽草酸/奎宁酸5-脱氢酶 |
| OsHEML | LOC_Os08g41990 | AGAACAAAGGGCAGATTGCTGCTG | TGTTTCGTCAAGTCACGGAGAGCA | 转氨酶 |
| OsHEMB | LOC_Os06g49110 | TGGCATTGTCAGGGAAGATGGAGT | CCAAAGCAGCACGTATTGCTCCAA | δ-氨基乙酰丙酸脱水酶,叶绿体前体 |
| GUN4 | LOC_Os11g16550 | AAGGGAAGGAGAGGCCAAAGTTCA | ACCATGACCAGCATCTCTGCATCA | 镁离子螯合酶H亚基结合蛋白 |
Table 2. Genes and primer sequences used in qRT-PCR.
| 基因 Gene | 基因登录号 Gene ID | 定量正向引物序列 Forward primer sequence | 定量反向引物序列 Reverse primer sequence | 功能 Function |
|---|---|---|---|---|
| NYC1 | LOC_Os01g12710 | CATGCAACACCAACAAAAGG | GACCATTCCAGGAGAAGCAG | 叶绿素b还原酶基因 |
| NOL | LOC_Os03g45194 | CCACGAAAGGTATAGGATATG | TCAAGTCAGTCACCGCAGAT | 叶绿素b还原酶基因 |
| NYC3 | LOC_Os06g24730 | TCTATCTAGGTGCCAAAGGC | ATTCTGGCACCTGCTGTTTC | α/β折叠水解酶家族蛋白 |
| PAO | LOC_Os03g05310 | AAGCCTCCGATGTTACCGAA | CGAGGGTTTCCAGAATTTGA | 脱镁叶绿酸a加氧酶,叶绿体前体 |
| SGR | LOC_Os09g36200 | GCAATGTCGCCAAATGACG | GCTCACCACACTCATTCCCTAAAG | 镁离子去螯合酶 |
| RCCR1 | LOC_Os10g25020 | GGATCGACGATTGATTTCATG | GTCGAGGCGTTCAGAAAGAT | 红色叶绿素分解代谢还原酶 |
| RCCR2 | LOC_Os10g25040 | TGGCGAGGGACAGGAAGGT | GGATGTGGTGGCGAGAGAAAC | 红色叶绿素分解代谢还原酶 |
| LchP2 | LOC_Os09g17740 | GAAGAAGATCAAGAACGGCC | TTGCCGGGGACGAAGTTGGT | 捕光叶绿素a/b结合蛋白基因 |
| PsbA | LOC_Os12g19580 | AGAGACGCGAAAGTACAAGC | AAGTTGCGGTCAATAAGGTA | 光合反应中心蛋白 |
| RpoC2 | LOC_Os04g16830 | ATGCATCGCAGGTACACCAA | CCCTCGCGTAAATTGCTTTG | DNA定向RNA聚合酶亚单位β |
| Rps15 | LOC_Os12g10580 | AGATACGGAGACTTGCTTCA | GCTCCCTAATATCCAACTGACT | 核酮糖二磷酸羧化酶大链前体 |
| V1 | LOC_Os03g45400 | AGAATCAGCGCGAGAAGAGAACCT | TACACCAGCTTTGGAGGAGCTGAA | 质体蛋白 |
| V2 | LOC_Os03g20460 | AGCAGATCCGTGATTACATGGCGA | TGCCTCTTCACTCTCTGCAACCAA | 鸟苷酸激酶 |
| YGL8 | LOC_Os01g17170 | TGGATCTAACATGACACGCACCCA | ACTGTAACGGCATTCTTCTCCGGT | 镁原卟啉IX单酯环化酶的催化亚基 |
| CAO1 | LOC_Os10g41780 | TTGGCTCAGTTAATGAGGGCAGAATCC | GGATGCGCACGTTGAGCATCTTTGTGG | 叶绿素a加氧酶 |
| PORA | LOC_Os04g58200 | ATGGCTCTCCAAGTTCAG | TGGCTCACGCTAAGGAAC | NADPH:原叶绿素酸酯氧化还原酶A |
| PORB | LOC_Os10g35370 | CCGCAAGGAGGGAGCGGTG | CCCTCTTGGTGCTAAGGCCG | 原叶绿素酸酯氧化还原酶B |
| CHLH | LOC_Os03g20700 | GCACGGGAACTTGGCGTTTCATTA | ACATGTCCTGGAGCTGCTTCTCAT | 镁离子螯合酶H亚基 |
| CHLD | LOC_Os03g59640 | TAGCACAGCTGTCAGAGTGGGTTT | TTGCCAGCCACCTCAAGTATCTCA | 镁离子螯合酶D亚基 |
| CHLI | LOC_Os03g36540 | AGGGATGCTGAACTCAGGGTGAAA | AAGTAGGACTCACGGAACGCCTTT | 镁离子螯合酶I亚基 |
| DVR | LOC_Os03g22780 | AGCCCAGGTTCATCAAGGT | TGATCACCCTCTCGAAGAACT | 联乙烯还原酶基因 |
| OsCHLM | LOC_Os06g04150 | GCTTCATCTCCACGCAGTTCTACT | GCAATGACGAATCGAAGACGCACA | 镁原卟啉O-甲基转移酶 |
| YGL1 | LOC_Os05g28200 | CCAGCCACTGATGAAAGCAGCAAT | AGAGCGCTAATACACTCGCGAACA | 叶绿素合成酶 |
| OsHEMA1 | LOC_Os10g35840 | GATGCAATCACTGCTGGAAAGCGT | CCATCTTGCCAGCACCAATCAACA | 莽草酸/奎宁酸5-脱氢酶 |
| OsHEML | LOC_Os08g41990 | AGAACAAAGGGCAGATTGCTGCTG | TGTTTCGTCAAGTCACGGAGAGCA | 转氨酶 |
| OsHEMB | LOC_Os06g49110 | TGGCATTGTCAGGGAAGATGGAGT | CCAAAGCAGCACGTATTGCTCCAA | δ-氨基乙酰丙酸脱水酶,叶绿体前体 |
| GUN4 | LOC_Os11g16550 | AAGGGAAGGAGAGGCCAAAGTTCA | ACCATGACCAGCATCTCTGCATCA | 镁离子螯合酶H亚基结合蛋白 |
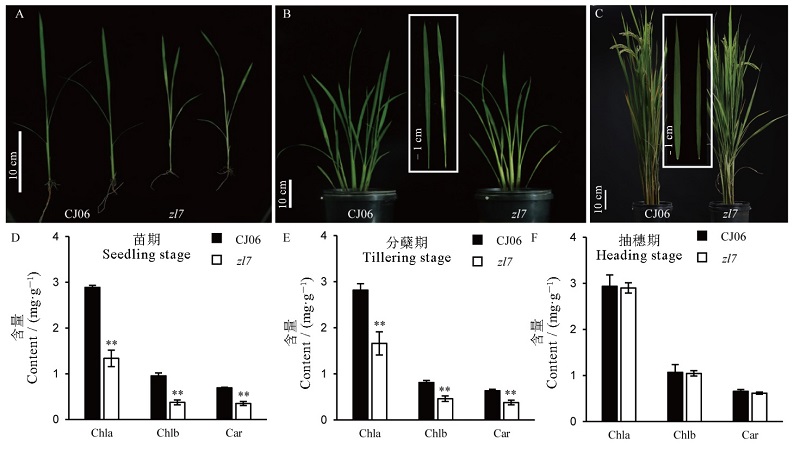
Fig. 1. Phenotypes of wild type CJ06 and zl7 at different growth stages. A, B and C are the phenotypes of wild-type Chunjiang06 and zl7 plants at seedling stage, tillering stage, and heading stage. The white box in figure B and C are enlarged leaves. D, E and F are chlorophyll contents at seedling stage, tillering stage and heading stage, respectively; Chla, Chlorophyll a; Chlb, Chlorophyll b; Car, Carotenoid; Means±SD(n=3); *,** indicates that there is significant difference between the wild type and the mutant at the level of 0.05 and 0.01 (t-test).
| 农艺性状 Agronomic trait | 野生型 Wild type | 突变体 Mutant zl7 |
|---|---|---|
| 株高 Plant height/cm | 91.2±4.0 | 79.5±2.7** |
| 分蘖 Tillering number | 10.0±1.2 | 8.0±0.8* |
| 穗长 Panicle length/cm | 22.5±1.3 | 20.2±0.8* |
| 一次枝梗 Primary rachis branches | 17.0±0.5 | 14.0±1.1* |
| 二次枝梗 Secondary rachis branches | 28.0±1.9 | 23.0±1.6* |
| 每穗粒数 No. of grains per panicle | 152.0±6.0 | 126.0±4.7** |
| 结实率Seed setting rate/% | 93.4±0.0 | 93.7±0.0 |
| 千粒重 1000-grain weight/g | 23.2±0.6 | 24.9±0.4 |
| 粒长 Grain length/mm | 7.35±0.40 | 7.67±0.63 |
| 粒宽 Grain width/mm | 3.08±0.23 | 3.17±0.24 |
Table 3. Comparison of main agronomic traits between wild type Chunjiang 06 and mutant zl7.
| 农艺性状 Agronomic trait | 野生型 Wild type | 突变体 Mutant zl7 |
|---|---|---|
| 株高 Plant height/cm | 91.2±4.0 | 79.5±2.7** |
| 分蘖 Tillering number | 10.0±1.2 | 8.0±0.8* |
| 穗长 Panicle length/cm | 22.5±1.3 | 20.2±0.8* |
| 一次枝梗 Primary rachis branches | 17.0±0.5 | 14.0±1.1* |
| 二次枝梗 Secondary rachis branches | 28.0±1.9 | 23.0±1.6* |
| 每穗粒数 No. of grains per panicle | 152.0±6.0 | 126.0±4.7** |
| 结实率Seed setting rate/% | 93.4±0.0 | 93.7±0.0 |
| 千粒重 1000-grain weight/g | 23.2±0.6 | 24.9±0.4 |
| 粒长 Grain length/mm | 7.35±0.40 | 7.67±0.63 |
| 粒宽 Grain width/mm | 3.08±0.23 | 3.17±0.24 |
| 材料 Material | 光合作用速率 Pn/ (μmol·m−2 s−1) | 气孔导度 Gs / (mol·m−2 s−1) | 胞间CO2浓度 Ci / (μmol·mol−1) | 蒸腾速率 Tr/ (mol·m−2 s−1) |
|---|---|---|---|---|
| 野生型WT | 26.41±0.34 | 0.06±0.01 | 442.34±24.64 | 2.94±0.08 |
| Mutant-g | 21.67±0.51 ** | 0.05±0.01 | 246.63±29.41** | 2.83±0.32 |
| Mutant-y | 13.21±0.47 ** | 0.04±0.01 | 133.69±7.49** | 2.34±0.17* |
Table 4. Photosynthetic parameters of the wild type CJ06 and the mutant zl7 at peak tillering stage.
| 材料 Material | 光合作用速率 Pn/ (μmol·m−2 s−1) | 气孔导度 Gs / (mol·m−2 s−1) | 胞间CO2浓度 Ci / (μmol·mol−1) | 蒸腾速率 Tr/ (mol·m−2 s−1) |
|---|---|---|---|---|
| 野生型WT | 26.41±0.34 | 0.06±0.01 | 442.34±24.64 | 2.94±0.08 |
| Mutant-g | 21.67±0.51 ** | 0.05±0.01 | 246.63±29.41** | 2.83±0.32 |
| Mutant-y | 13.21±0.47 ** | 0.04±0.01 | 133.69±7.49** | 2.34±0.17* |
| 材料 Material | 光合作用速率 Pn/ (μmol·m−2·s−1) | 气孔导度 Gs / (mol·m−2·s−1) | 胞间CO2浓度 Ci / (μmol·mol−1) | 蒸腾速率 Tr/ (mol·m−2·s−1) |
|---|---|---|---|---|
| WT | 16.06±0.56 | 0.15±0.02 | 204.24±17.42 | 3.29±0.30 |
| Mutant-g | 18.34±0.64** | 0.28±0.02* | 278.66±9.48** | 4.79±0.26** |
| Mutant-y | 9.93±1.41** | 0.13±0.03 | 262.71±15.41** | 2.71±0.58 |
Table 5. Photosynthetic parameters of wild type CJ06 and mutant zl7 at heading stage.
| 材料 Material | 光合作用速率 Pn/ (μmol·m−2·s−1) | 气孔导度 Gs / (mol·m−2·s−1) | 胞间CO2浓度 Ci / (μmol·mol−1) | 蒸腾速率 Tr/ (mol·m−2·s−1) |
|---|---|---|---|---|
| WT | 16.06±0.56 | 0.15±0.02 | 204.24±17.42 | 3.29±0.30 |
| Mutant-g | 18.34±0.64** | 0.28±0.02* | 278.66±9.48** | 4.79±0.26** |
| Mutant-y | 9.93±1.41** | 0.13±0.03 | 262.71±15.41** | 2.71±0.58 |
| 杂交组合 Hybrid combination | F1 | F2 | χ2 (3:1) | ||
|---|---|---|---|---|---|
| 正常表型 Wild-type phenotype | 突变表型 zl7 phenotype | 正常表型 Wild-type phenotype | 突变表型 zl7 phenotype | ||
| zl7/93-11 | 13 | 0 | 2065 | 655 | 1.225 |
| 93-11/ zl7 | 8 | 0 | 822 | 271 | 0.025 |
Table 6. Genetic analysis of the mutant zl7.
| 杂交组合 Hybrid combination | F1 | F2 | χ2 (3:1) | ||
|---|---|---|---|---|---|
| 正常表型 Wild-type phenotype | 突变表型 zl7 phenotype | 正常表型 Wild-type phenotype | 突变表型 zl7 phenotype | ||
| zl7/93-11 | 13 | 0 | 2065 | 655 | 1.225 |
| 93-11/ zl7 | 8 | 0 | 822 | 271 | 0.025 |
| 预测基因 Predictive gene | 基因功能注释 Gene function annotation |
|---|---|
| ORF1 | 转座子蛋白 |
| ORF2 | 表达蛋白 |
| ORF3 | ζ-胡萝卜素脱氢酶 |
| ORF4 | 表达蛋白 |
| ORF5 | 细胞色素b-c1复合亚单位7 |
| ORF6 | 表达蛋白 |
| ORF7 | 核苷酸结合蛋白 |
| ORF8 | 羧基末端肽酶 |
| ORF9 | 表达蛋白 |
| ORF10 | 肽酶,M24家族蛋白 |
| ORF11 | 细胞周期蛋白相关蛋白 |
| ORF12 | 表达蛋白 |
Table 7. Gene function annotation of ORFs.
| 预测基因 Predictive gene | 基因功能注释 Gene function annotation |
|---|---|
| ORF1 | 转座子蛋白 |
| ORF2 | 表达蛋白 |
| ORF3 | ζ-胡萝卜素脱氢酶 |
| ORF4 | 表达蛋白 |
| ORF5 | 细胞色素b-c1复合亚单位7 |
| ORF6 | 表达蛋白 |
| ORF7 | 核苷酸结合蛋白 |
| ORF8 | 羧基末端肽酶 |
| ORF9 | 表达蛋白 |
| ORF10 | 肽酶,M24家族蛋白 |
| ORF11 | 细胞周期蛋白相关蛋白 |
| ORF12 | 表达蛋白 |

Fig. 4. Expression analysis of chloroplast development related genes of the wild type Chunjiang 06(CJ06) and its mutant zl7. Mean±SD(n=3). *, ** indicates that there is significant difference between wild type(CJ06) and mutant at the level of 0.05 and 0.01 respectively (t-test)
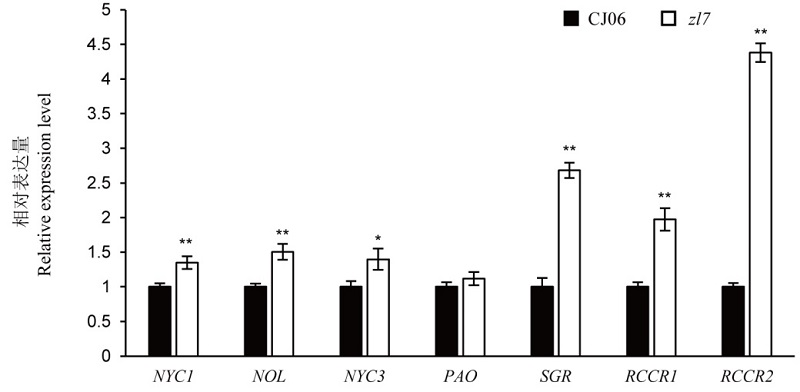
Fig. 6. Expression analysis of chlorophyll degradation related genes of the wild type Chunjiang 06(CJ06) and its mutant zl7. Mean±SD (n=3). *, ** indicate that there is significant difference between the wild type and the mutant at the level of 0.05 and 0.01 respectively (t-test).
| [1] | Hao J, Wang D, Wu Y, Huang K, Duan P, Li N, Xu R, Zeng D, Dong G, Zhang B, Zhang L, Inze D, Qian Q, Li Y. The GW2-WG1-OsbZIP47 pathway controls grain size and weight in rice[J]. Molecular Plant, 2021, 14(8): 1266-1280. |
| [2] | Tian Z, Wang J W, Li J, Han B. Designing future crops: Challenges and strategies for sustainable agriculture[J]. Plant Journal, 2021, 105(5): 1165-1178. |
| [3] | Fromme P, Melkozernov A, Jordan P, Krauss N. Structure and function of photosystem: I. Interaction with its soluble electron carriers and external antenna systems[J]. FEBS Letters, 2003, 555(1): 40-44. |
| [4] | Zhen X H, Xu J G, Shen W J, Zhang X J, Zhang Q J, Lu C G, Chen G X, Gao Z P. Photosynthetic characteristics of flag leaves in rice white stripe mutant 6001 during senescence process[J]. Rice Science, 2014, 21(6): 335-342. |
| [5] | Yang Y, Xu J, Huang L, Leng Y, Dai L, Rao Y, Chen L, Wang Y, Tu Z, Hu J, Ren D, Zhang G, Zhu L, Guo L, Qian Q, Zeng D. PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice[J]. Journal of Experimental Botany, 2016, 67(5): 1297-1310. |
| [6] | Cao W, Zhang H, Zhou Y, Zhao J, Lu S, Wang X, Chen X, Yuan L, Guan H, Wang G, Shen W, De Vleesschauwer D, Li Z, Shi X, Gu J, Guo M, Feng Z, Chen Z, Zhang Y, Pan X, Liu W, Liang G, Yan C, Hu K, Liu Q, Zuo S. Suppressing chlorophyll degradation by silencing OsNYC3 improves rice resistance to Rhizoctonia solani, the causal agent of sheath blight[J]. Plant Biotechnology Journal, 2022, 20(2): 335-349. |
| [7] | Yoo S C, Cho S H, Sugimoto H, Li J, Kusumi K, Koh H J, Iba K, Paek N C. Rice virescent3 and stripe1encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development[J]. Plant Physiology, 2009, 150(1): 388-401. |
| [8] | Zhu X, Ze M, Yin J, Chen M, Wang M, Zhang X, Deng R, Li Y, Liao H, Wang L, Tu B, Song L, He M, Li S, Wang W M, Chen X, Wang J, Li W. A phosphofructokinase B-type carbohydrate kinase family protein, PFKB1, is essential for chloroplast development at early seedling stage in rice[J]. Plant Science, 2020, 290: 110295. |
| [9] | 杨颜榕, 黄纤纤, 赵亚男, 汤佳玉, 刘喜. 水稻叶色基因克隆与分子机制研究进展[J]. 植物遗传资源学报, 2020, 21(4): 794-803. |
| Yang Y R, Huang X X, Zhao Y N, Tang J Y, Liu X. Advances on gene isolation and molecular mechanism of rice leaf color genes[J]. Journal of Plant Genetic Resources, 2020, 21(4): 794-803. (in Chinese with English abstract) | |
| [10] | Kusumi K, Sakata C, Nakamura T, Kawasaki S, Yoshimura AIba K. A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions[J]. Plant Journal, 2011, 68(6):1039-1050. |
| [11] | Wang P, Gao J, Wan C, Zhang F, Xu Z, Huang X, Sun X, Deng X. Divinyl chlorophyll(ide) a can be converted to monovinyl chlorophyll(ide) a by a divinyl reductase in rice[J]. Plant Physiology, 2010, 153(3): 994-1003. |
| [12] | Yang Y L, Xu J, Rao Y C, Zeng Y J, Liu H J, Zheng T T, Zhang G H, Hu J, Guo L B, Qian Q, Zeng D L, Shi Q H. Cloning and functional analysis of pale-green leaf (PGL10) in rice (Oryza sativa L.)[J]. Plant Growth Regulation, 2016, 78(1): 69-77. |
| [13] | Tang J, Zhang W, Wen K, Chen G, Sun J, Tian Y, Tang W, Yu J, An H, Wu T, Kong F, Terzaghi W, Wang C, Wan J. OsPPR6, a pentatricopeptide repeat protein involved in editing and splicing chloroplast RNA, is required for chloroplast biogenesis in rice[J]. Plant Molecular Biology, 2017, 95(4-5): 345-357. |
| [14] | Shin D, Lee S, Kim T H, Lee J H, Park J, Lee J, Lee J Y, Cho L H, Choi J Y, Lee W, Park J H, Lee D W, Ito H, Kim D H, Tanaka A, Cho J H, Song Y C, Hwang D, Purugganan M D, Jeon J S, An G, Nam H G. Natural variations at the Stay-Green gene promoter control lifespan and yield in rice cultivars[J]. Nature Communications, 2020, 11(1): 2819. https://doi.org/10.1038/s41467-020-16573-2. |
| [15] | Liu Z, Wang Z, Gu H, You J, Hu M, Zhang Y, Zhu Z, Wang Y, Liu S, Chen L, Liu X, Tian Y, Zhou S, Jiang L, Liu L, Wan J. Identification and phenotypic characterization of ZEBRA LEAF16 encoding a beta-hydroxyacyl-ACP dehydratase in rice[J]. Frontiers in Plant Science, 2018, 9: 782. |
| [16] | Feng P, Shi J, Zhang T, Zhong Y, Zhang L, Yu G, Zhang T, Zhu X, Xing Y, Yin W, Sang X, Ling Y, Zhang C, Yang Z, He G, Wang N. Zebra leaf 15, a receptor-like protein kinase involved in moderate low temperature signaling pathway in rice[J]. Rice (N Y), 2019, 12(1): 83. |
| [17] | Li J, Pandeya D, Nath K, Zulfugarov I S, Yoo S C, Zhang H, Yoo J H, Cho S H, Koh H J, Kim D S, Seo H S, Kang B C, Lee C H, Paek N C. ZEBRA-NECROSIS, a thylakoid-bound protein, is critical for the photoprotection of developing chloroplasts during early leaf development[J]. Plant Journal, 2010, 62(4): 713-725. |
| [18] | Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method[J]. Methods, 2001, 25(4): 402-408. |
| [19] | Sugimoto H, Kusumi K, Noguchi K, Yano M, Yoshimura A, Iba K. The rice nuclear gene, VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria[J]. Plant Journal, 2007, 52(3): 512-527. |
| [20] | Sakuraba Y, Rahman M L, Cho S H, Kim Y S, Koh H J, Yoo S C, Paek N C. The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions[J]. Plant Journal, 2013, 74(1): 122-133. |
| [21] | Jiang H, Li M, Liang N, Yan H, Wei Y, Xu X, Liu J, Xu Z, Chen F, Wu G. Molecular cloning and function analysis of the stay green gene in rice[J]. Plant Journal, 2007, 52(2): 197-209. |
| [22] | Shin D, Lee S, Kim T H, Lee J H, Park J, Lee J, Lee J Y, Cho L H, Choi J Y, Lee W, Park J H, Lee D W, Ito H, Kim D H, Tanaka A, Cho J H, Song Y C, Hwang D, Purugganan M D, Jeon J S, An G, Nam H G. Natural variations at the Stay-Green gene promoter control lifespan and yield in rice cultivars[J]. Nature Communications, 2020, 11(1): 2819. |
| [23] | 方希林, 王悦, 朱敏敏, 邓跃军, 张晶李恩宇. 水稻叶色突变基因的遗传研究进展[J]. 北方水稻, 2016, 46(6): 4-8. |
| Fang X L, Wang Y, Zhu M M, Deng Y J, Zhang J, Li E Y. Research progress of rice leaf color mutation genetic analysis[J]. North Rice, 2016, 46(6): 4-8. (in Chinese with English abstract) | |
| [24] | Beale S I. Green genes gleaned[J]. Trends in Plant Science, 2005, 10(7): 309-312. |
| [25] | 李佳佳, 于旭东, 蔡泽坪, 吴繁花, 罗佳佳, 郑李婷, 楚文清. 高等植物叶绿素生物合成研究进展[J]. 分子植物育种, 2019, 17(18): 6013-6019. |
| Li J J, Yu X D, Cai Z P, Wu F H, Luo J J, Zheng L T, Chu W Q. An overview of chlorophyll biosynthesis in higher plants[J]. Molecular Plant Breeding, 2019, 17(18): 6013-6019. | |
| [26] | 赵绍路, 刘凯, 宛柏杰, 朱静雯, 刘艳艳, 唐红生, 严国红, 孙明法. 水稻叶色突变研究进展[J]. 大麦与谷类科学, 2018, 35(6): 1-6. |
| Zhao S L, Liu K, Wan B J, Zhu J W, Liu Y Y, Tang H S, Yan G H, Sun M F. Advances in research on rice leaf color mutants[J]. Barley and Cereal Sciences, 2018, 35(6): 1-6. (in Chinese with English abstract) | |
| [27] | Cazzonelli C I, Pogson B J. Source to sink: regulation of carotenoid biosynthesis in plants[J]. Trends in Plant Science, 2010, 15(5): 266-274. |
| [28] | 陆晨飞, 刘钰婷. 类胡萝卜素代谢调控与植物颜色变异[J]. 北方园艺, 2016, 16: 193-199. |
| Lu C F, Liu Y T. Plant color mutants and the regulation of carotenoids metabolism[J]. Northern Horticulture, 2016, 16: 193-199. (in Chinese with English abstract) | |
| [29] | Kusumi K, Komori H, Satoh H, Iba K. Characterization of a zebra mutant of rice with increased susceptibility to light stress[J]. Plant Cell Physiology, 2000, 41(2): 158-164. |
| [30] | Sakuraba Y. Light-mediated regulation of leaf senescence[J]. International Journal of Molecular Medicine, 2021, 22(7). |
| [31] | 张天雨, 周春雷, 刘喜, 孙爱伶, 曹鹏辉, Nguyen T, 田云录, 翟虎渠, 江玲. 一个水稻温敏黄化突变体的表型分析和基因定位[J]. 作物学报, 2017, 43(10): 1426-1433. |
| Zhang T Y, Zhou C L, Liu X, Sun A L, Cao P H, Nguyen T, Tian Y L, Zhai H Q, Jiang L. Phenotypes and gene mapping of a thermo-sensitive yellow leaf mutant of rice[J]. Acta Agronomica Sinica, 2017, 43(10): 1426-1433. (in Chinese with English abstract) | |
| [32] | 王威, 张联合, 李华, 张志华, 胡斌, 储成才. 水稻营养吸收和转运的分子机制研究进展[J]. 中国科学: 生命科学, 2015, 45(6):569-590. |
| Wang W, Zhang L H, Li H, Zhang Z H, Hu B, Chu C C. Recent progress in molecular dissection of nutrient uptake and transport in rice[J]. Scientia Sinica: Vitae, 2015, 45(6): 569-590. | |
| [33] | Long J R, Ma G H, Wan Y Z, Song C F, Sun J, Qin R J. Effects of nitrogen fertilizer level on chlorophyll fluorescence characteristics in flag leaf of super hybrid rice at late growth stage[J]. Rice Science, 2013, 20(3): 220-228. |
| [34] | Carol P, Stevenson D, Bisanz C, Breitenbach J, Sandmann G, Mache R, Coupland G, Kuntz M. Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation[J]. Plant Cell, 1999, 11(1): 57-68. |
| [35] | Chen M, Choi Y, Voytas D F, Rodermel S. Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease[J]. Plant Journal, 2000, 22(4): 303-313. |
| [36] | Naested H, Holm A, Jenkins T, Nielsen H B, Harris C A, Beale M H, Andersen M, Mant A, Scheller H, Camara B, Mattsson O, Mundy J. Arabidopsis VARIEGATED 3 encodes a chloroplast-targeted, zinc-finger protein required for chloroplast and palisade cell development[J]. Journal of Cell Science, 2004, 117(Pt 20): 4807-4818. |
| [37] | Wang Y, Shang L, Yu H, Zeng L, Hu J, Ni S, Rao Y, Li S, Chu J, Meng X, Wang L, Hu P, Yan J, Kang S, Qu M, Lin H, Wang T, Wang Q, Hu X, Chen H, Wang B, Gao Z, Guo L, Zeng D, Zhu X, Xiong G, Li J, Qian Q. A strigolactone biosynthesis gene contributed to the green revolution in rice[J]. Molecular Plant, 2020, 13(6): 923-932. |
| [38] | Zou J, Zhang S, Zhang W, Li G, Chen Z, Zhai W, Zhao X, Pan X, Xie Q, Zhu L. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds[J]. Plant Journal, 2006, 48(5): 687-698. |
| [39] | Wang Y, Wang C, Zheng M, Lyu J, Xu Y, Li X, Niu M, Long W, Wang D, Wang H, Terzaghi W, Wang Y, Wan J. WHITE PANICLE1, a val-tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development[J]. Plant Physiology, 2016, 170(4): 2110-2123. |
| [40] | Wang Z W, Lü J, Xie S Z, Zhang Y, Qiu Z N, Chen P, Cui Y T, Niu Y F, Hu S K, Jiang H Z, Ge S Z, Trinh H, Lei K R, Bai W Q, Zhang Y, Guo L B, Ren D Y. OsSLA4 encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling growth in rice[J]. Plant Growth Regulation, 2018, 84(2): 249-260. |
| [41] | Ma X, Ma J, Zhai H, Xin P, Chu J, Qiao Y, Han L. CHR729 is a CHD3 protein that controls seedling development in rice[J]. PLoS One, 2015, 10(9): e0138934. |
| [42] | Bang W Y, Chen J, Jeong I S, Kim S W, Kim C W, Jung H S, Lee K H, Kweon H S, Yoko I, Shiina T, Bahk J D. Functional characterization of ObgC in ribosome biogenesis during chloroplast development[J]. Plant Journal, 2012, 71(1): 122-134. |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||