
Chinese Journal OF Rice Science ›› 2021, Vol. 35 ›› Issue (6): 543-553.DOI: 10.16819/j.1001-7216.2021.201211
• 研究报告 • Previous Articles Next Articles
Ke GONG1, Pao XUE1, Xiaoxia WEN1, Feifei LIAO1, Bin SUN2, Zequn PENG1, Shihua CHENG1, Liyong CAO1, Yingxin ZHANG1, Weixun WU1, Lianping SUN1,*( ), Xiaodeng ZHAN1,*(
), Xiaodeng ZHAN1,*( )
)
Received:2020-12-15
Revised:2021-02-09
Online:2021-11-10
Published:2021-11-10
Contact:
Lianping SUN, Xiaodeng ZHAN
龚柯1, 薛炮1, 温小霞1, 廖飞飞1, 孙滨2, 彭泽群1, 程式华1, 曹立勇1, 张迎信1, 吴玮勋1, 孙廉平1,*( ), 占小登1,*(
), 占小登1,*( )
)
通讯作者:
孙廉平,占小登
基金资助:Ke GONG, Pao XUE, Xiaoxia WEN, Feifei LIAO, Bin SUN, Zequn PENG, Shihua CHENG, Liyong CAO, Yingxin ZHANG, Weixun WU, Lianping SUN, Xiaodeng ZHAN. Distribution of Grain Shape Related Genes in Rice Big Grain Germplasm BG1 and Elite Restorer Line Huazhan and Development of Relevant Functional Markers[J]. Chinese Journal OF Rice Science, 2021, 35(6): 543-553.
龚柯, 薛炮, 温小霞, 廖飞飞, 孙滨, 彭泽群, 程式华, 曹立勇, 张迎信, 吴玮勋, 孙廉平, 占小登. 水稻特大粒种质BG1和优质恢复系华占的粒形基因研究及相关功能标记开发[J]. 中国水稻科学, 2021, 35(6): 543-553.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2021.201211
| 引物名称 Primer name | 前引物 Forward primer | 后引物 Reverse primer | 产物大小 Product size/bp |
|---|---|---|---|
| GS3-seq | ACTGTATGCTCAAAGCATCT | TGCTTTACTTTTTTTTTTGC | 1593 |
| GL3.1-seq | GCCACTCATGCACCATAACTAC | ACCTCCTCGTAGACCTCCATAA | 435 |
| LGY3-cds-1 | GCTGAAGCGGATCGAGAACAA | GCCGGATGGGATGTGTTCATT | 744 |
| LGY3-YZ | ACTTGTTACCACATCCAAAACGG | AATCCTGACAATAATTCGCCCA | 522 |
| GW2-cds-1 | CAGGGACATCGACCAGAAGAA | TACAACCATGCCAACCCTTG | 1199 |
| GW5-indel-3 | TCCATGCAGTACTTGGTTTCC | AGATCTGAGCGGTAACTTTCTC | 938/2150 |
| GW8-indel-2 | AGCTACAGAATCCAGAAACAAACCA | CTGGCCCCTTCCCCTTGG | 611 |
| GS9-seq | GTTCCAACGCCAACAGCAC | CTTCGCGGTAGCGATCCAAT | 2601 |
| TGW3-cds-1 | CCGGCTAGTTCGGTTGAATG | AGACGTTCGATGGCTTCTGG | 1153/1233 |
| TGW6-cds-1 | CCACAGCCACAACGAGAATG | CGACTCCGACATACGGCAAT | 947 |
Table 1 Sequencing primers used in the study.
| 引物名称 Primer name | 前引物 Forward primer | 后引物 Reverse primer | 产物大小 Product size/bp |
|---|---|---|---|
| GS3-seq | ACTGTATGCTCAAAGCATCT | TGCTTTACTTTTTTTTTTGC | 1593 |
| GL3.1-seq | GCCACTCATGCACCATAACTAC | ACCTCCTCGTAGACCTCCATAA | 435 |
| LGY3-cds-1 | GCTGAAGCGGATCGAGAACAA | GCCGGATGGGATGTGTTCATT | 744 |
| LGY3-YZ | ACTTGTTACCACATCCAAAACGG | AATCCTGACAATAATTCGCCCA | 522 |
| GW2-cds-1 | CAGGGACATCGACCAGAAGAA | TACAACCATGCCAACCCTTG | 1199 |
| GW5-indel-3 | TCCATGCAGTACTTGGTTTCC | AGATCTGAGCGGTAACTTTCTC | 938/2150 |
| GW8-indel-2 | AGCTACAGAATCCAGAAACAAACCA | CTGGCCCCTTCCCCTTGG | 611 |
| GS9-seq | GTTCCAACGCCAACAGCAC | CTTCGCGGTAGCGATCCAAT | 2601 |
| TGW3-cds-1 | CCGGCTAGTTCGGTTGAATG | AGACGTTCGATGGCTTCTGG | 1153/1233 |
| TGW6-cds-1 | CCACAGCCACAACGAGAATG | CGACTCCGACATACGGCAAT | 947 |
| 种质 | 千粒重 | 增幅 | 长宽比 | 增幅 | 粒长 | 增幅 | 粒宽 | 增幅 |
|---|---|---|---|---|---|---|---|---|
| Germplasm | TGW/g | Increase/% | GL/GW | Increase/% | GL/mm | Increase/% | GW/mm | Increase/% |
| 日本晴Nipponbare | 26.11 ± 0.33 | 2.19 ± 0.01 | 6.98 ± 0.01 | 3.19 ± 0.01 | ||||
| BG1 | 62.97 ± 0.01 | 141.17 | 2.85 ± 0.04 | 30.14 | 12.18 ± 0.04 | 74.45 | 4.27 ± 0.06 | 33.86 |
| 华占Huazhan | 18.44 ± 0.13 | -29.38 | 3.85 ± 0.03 | 75.80 | 8.28 ± 0.08 | 16.62 | 2.15 ± 0.04 | -32.60 |
Table 2 Grain shape characters of Nipponbare, BG1 and HZ.
| 种质 | 千粒重 | 增幅 | 长宽比 | 增幅 | 粒长 | 增幅 | 粒宽 | 增幅 |
|---|---|---|---|---|---|---|---|---|
| Germplasm | TGW/g | Increase/% | GL/GW | Increase/% | GL/mm | Increase/% | GW/mm | Increase/% |
| 日本晴Nipponbare | 26.11 ± 0.33 | 2.19 ± 0.01 | 6.98 ± 0.01 | 3.19 ± 0.01 | ||||
| BG1 | 62.97 ± 0.01 | 141.17 | 2.85 ± 0.04 | 30.14 | 12.18 ± 0.04 | 74.45 | 4.27 ± 0.06 | 33.86 |
| 华占Huazhan | 18.44 ± 0.13 | -29.38 | 3.85 ± 0.03 | 75.80 | 8.28 ± 0.08 | 16.62 | 2.15 ± 0.04 | -32.60 |
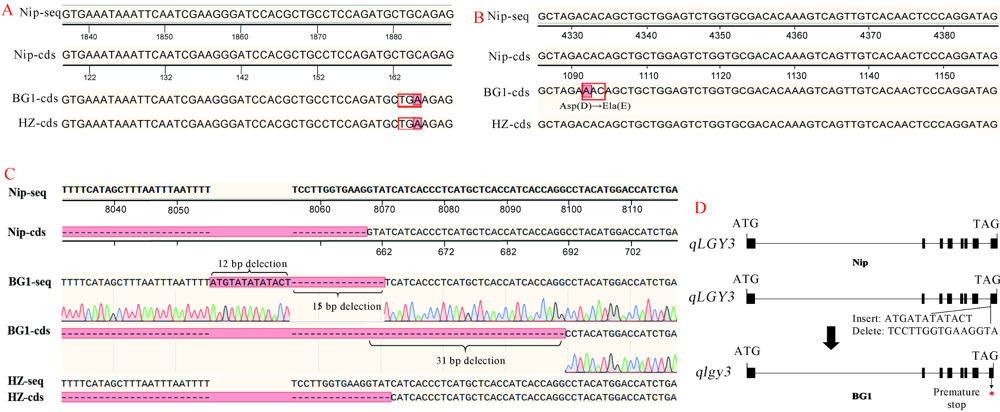
Fig. 2. Detection results of genes for grain length. A, Sequence of GS3; B, Sequence of qGL3; C, Sequence of qLGY3; D, Exons structure of qLGY3 in BG1. Nip-seq, Genomic sequence of Nipponbare; Nip-cds, CDS sequence of Nipponbare; BG1-seq, Genome sequence of BG1; BG1-cds, CDS sequence of BG1; HZ-seq, Genome sequence of Huazhan; HZ-cds, CDS sequence of Huazhan. Nip, Nipponbare. BG1, Big Grain 1. HZ, Huazhan.
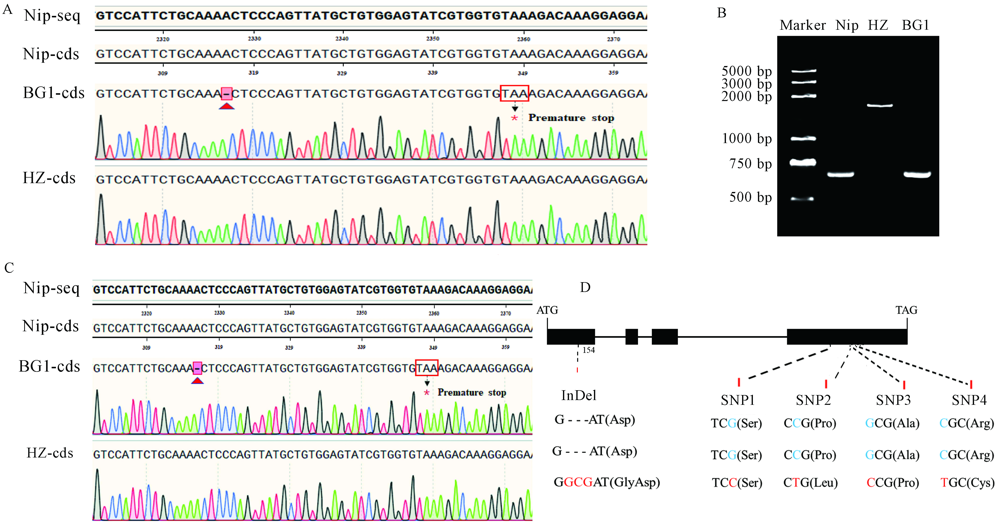
Fig. 3. Detection results of genes for grain width. A, Sequence of GW2; B, Agarose gel test of GW5; C, Sequence of GW8; D, Sequence of GS9. Nip-seq, Genomic sequence of Nipponbare; Nip-cds, Sequence of Nipponbare; BG1-seq, Genome sequence of BG1; BG1-cds, CDS sequence of BG1; HZ-seq, Genome sequence of Huazhan. HZ-cds, CDS sequence of Huazhan. Nip, Nipponbare. HZ, Huazhan.
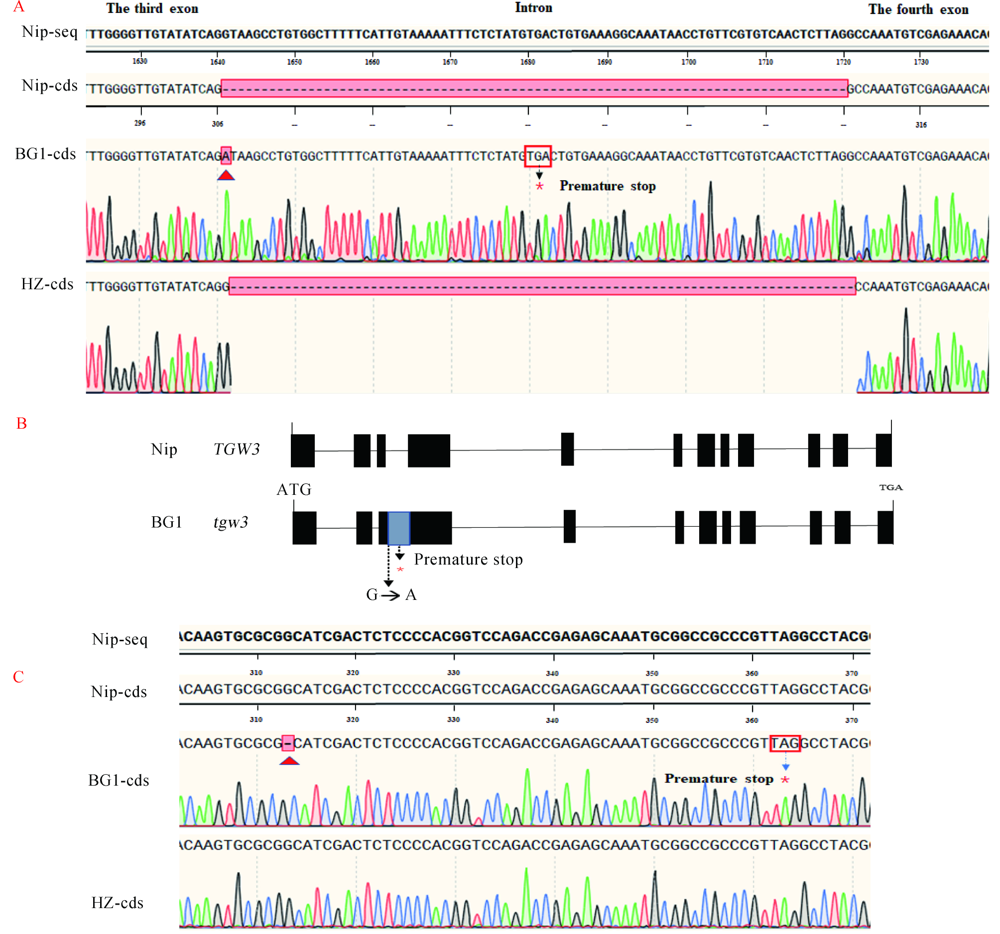
Fig. 4. Detection results of genes for 1000-grain weight. A, CDS sequence of TGW3. B, Exons structure of TGW3. C, CDS sequence of TGW6. Nip-seq, Genomic sequence of Nipponbare. Nip-cds, CDS sequence of Nipponbare. BG1-cds, CDS sequence of BG1. HZ-cds, CDS sequence of Huazhan. Nip, Nipponbare.
| 材料Material | GS3 | qGL3 | qLGY3 | GW2 | GW8 | GW5 | GS9 | TGW3 | TGW6 |
|---|---|---|---|---|---|---|---|---|---|
| 日本晴Nipponbare | - | - | - | - | gw8 | gw5 | - | - | - |
| BG1 | gs3 | qgl3 | qlgy3 | gw2 | - | gw5 | - | tgw3 | tgw6 |
| 华占Huazhan | gs3 | - | - | - | - | - | - | - | - |
Table 3 Genotypes of grain-shape-related genes in Nipponbare, BG1 and Huazhan.
| 材料Material | GS3 | qGL3 | qLGY3 | GW2 | GW8 | GW5 | GS9 | TGW3 | TGW6 |
|---|---|---|---|---|---|---|---|---|---|
| 日本晴Nipponbare | - | - | - | - | gw8 | gw5 | - | - | - |
| BG1 | gs3 | qgl3 | qlgy3 | gw2 | - | gw5 | - | tgw3 | tgw6 |
| 华占Huazhan | gs3 | - | - | - | - | - | - | - | - |
| 引物名称 Primer name | 前引物 Forward primer | 后引物 Reverse primer | 内切酶 Enzyme | 产物长度 Product size/bp |
|---|---|---|---|---|
| GS3-SNP | GGATCCACGCTGCCTCCAGATGGTG | TTGCCAAGGTTTTATAATCAATGGT | Hph Ⅰ | 225/203 |
| TGW3-SNP | CTACGATCCGTGGCCGAAA | TTACAATGAAAAAGCCACAGGCGTA | Rsa Ⅰ | 206/185 |
| qGL3-SNP | CGATTCTATCTGGTTCAGTGGTAGA | AAACAGGTTTTCTTACCTCCTCGT | Acc Ⅰ | 212/192/170 |
| GW2-InDel | CAACTCACACTGCTCAGCCTACA | ATACTCCACAGCATAACTGGGAGTCT | Pst Ⅰ | 133/99 |
| TGW6-InDel | AGCCCCAGCTACACGAAAAACAAGTCCGCG | CCCATTGGTGAAGCGAAGTG | Sac Ⅱ | 221/196 |
| LGY3-InDel | GGGCCTAATTTTGTCTTTGTTATTC | GTCTGCTGCTTCATTGCTCA | Sty Ⅰ | 162/85/77 |
| GW8-InDel | GCGTCAACACACAGCTCAAG | CGGCATCTTGAGATCCCACTC | 89/79 | |
| GW5-InDel3 | TCCATGCAGTACTTGGTTTCC | AGATCTGAGCGGTAACTTTCTC | 2150/938 |
Table 4 Function marker information of major grain shape genes.
| 引物名称 Primer name | 前引物 Forward primer | 后引物 Reverse primer | 内切酶 Enzyme | 产物长度 Product size/bp |
|---|---|---|---|---|
| GS3-SNP | GGATCCACGCTGCCTCCAGATGGTG | TTGCCAAGGTTTTATAATCAATGGT | Hph Ⅰ | 225/203 |
| TGW3-SNP | CTACGATCCGTGGCCGAAA | TTACAATGAAAAAGCCACAGGCGTA | Rsa Ⅰ | 206/185 |
| qGL3-SNP | CGATTCTATCTGGTTCAGTGGTAGA | AAACAGGTTTTCTTACCTCCTCGT | Acc Ⅰ | 212/192/170 |
| GW2-InDel | CAACTCACACTGCTCAGCCTACA | ATACTCCACAGCATAACTGGGAGTCT | Pst Ⅰ | 133/99 |
| TGW6-InDel | AGCCCCAGCTACACGAAAAACAAGTCCGCG | CCCATTGGTGAAGCGAAGTG | Sac Ⅱ | 221/196 |
| LGY3-InDel | GGGCCTAATTTTGTCTTTGTTATTC | GTCTGCTGCTTCATTGCTCA | Sty Ⅰ | 162/85/77 |
| GW8-InDel | GCGTCAACACACAGCTCAAG | CGGCATCTTGAGATCCCACTC | 89/79 | |
| GW5-InDel3 | TCCATGCAGTACTTGGTTTCC | AGATCTGAGCGGTAACTTTCTC | 2150/938 |
| 种质名称 Germplasm | 粒型特征 Grain morphology | 基因分型 Genotype | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGW | GL/GW | GL | GW | GS3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | GW5 | |||
| 日本晴 | 25.39±0.20 | 2.20±0.01 | 7.01±0.02 | 3.20±0.01 | GS3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | gw8 | gw5 | ||
| 9311 | 28.47±0.22 | 3.58±0.01 | 9.07±0.03 | 2.55±0.01 | gs3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | GW5/gw5 | ||
| 中恢9308 | 20.15±0.24 | 3.08±0.01 | 7.70±0.03 | 2.51±0.01 | GS3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | gw5 | ||
| 中恢8015 | 37.03±0.33 | 3.29±0.01 | 10.03±0.01 | 3.05±0.01 | gs3 | TGW3 | qGL3 | GW2 | tgw6 | qLGY3 | GW8 | GW5 | ||
| 长粒粳 | 29.72±0.15 | 3.59±0.01 | 9.51±0.01 | 2.66±0.01 | gs3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | gw8 | GW5/gw5 | ||
| BG1 | 62.97±0.01 | 2.85±0.04 | 12.18±0.04 | 4.27±0.06 | gs3 | tgw3 | qgl3 | gw2 | tgw6 | qlgy3 | GW8 | gw5 | ||
| IR6 | 23.23±0.29 | 4.06±0.02 | 9.20±0.04 | 2.27±0.01 | gs3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | GW5/gw5 | ||
| 华占 | 19.03±0.38 | 3.88±0.02 | 8.29±0.03 | 2.15±0.01 | gs3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | GW5 | ||
| 中花11 | 23.94±0.21 | 2.19±0.01 | 6.92±0.01 | 3.17±0.02 | GS3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | gw8 | gw5 | ||
| 02428 | 23.86±0.20 | 2.32±0.02 | 6.87±0.01 | 2.98±0.02 | GS3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | gw8 | gw5 | ||
| NJ11 | 25.34±0.13 | 2.59±0.01 | 7.49±0.01 | 2.90±0.01 | GS3 | TGW3 | qGL3 | GW2 | tgw6 | qLGY3 | GW8 | gw5 | ||
| 玉针香 | 27.17±0.07 | 5.63±0.02 | 11.65±0.04 | 2.08±0.01 | gs3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | GW5/gw5 | ||
| IR24 | 24.49±0.09 | 3.63±0.02 | 8.81±0.04 | 2.44±0.02 | gs3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | GW5/gw5 | ||
| IR26 | 23.93±0.10 | 3.6±0.01 | 8.76±0.03 | 2.44±0.01 | gs3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | GW5/gw5 | ||
| IR64 | 24.57±0.12 | 4.01±0.02 | 9.07±0.03 | 2.27±0.01 | gs3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | GW5/gw5 | ||
Table 5 Grain morphology and genotyping of 15 germplasm.
| 种质名称 Germplasm | 粒型特征 Grain morphology | 基因分型 Genotype | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGW | GL/GW | GL | GW | GS3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | GW5 | |||
| 日本晴 | 25.39±0.20 | 2.20±0.01 | 7.01±0.02 | 3.20±0.01 | GS3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | gw8 | gw5 | ||
| 9311 | 28.47±0.22 | 3.58±0.01 | 9.07±0.03 | 2.55±0.01 | gs3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | GW5/gw5 | ||
| 中恢9308 | 20.15±0.24 | 3.08±0.01 | 7.70±0.03 | 2.51±0.01 | GS3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | gw5 | ||
| 中恢8015 | 37.03±0.33 | 3.29±0.01 | 10.03±0.01 | 3.05±0.01 | gs3 | TGW3 | qGL3 | GW2 | tgw6 | qLGY3 | GW8 | GW5 | ||
| 长粒粳 | 29.72±0.15 | 3.59±0.01 | 9.51±0.01 | 2.66±0.01 | gs3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | gw8 | GW5/gw5 | ||
| BG1 | 62.97±0.01 | 2.85±0.04 | 12.18±0.04 | 4.27±0.06 | gs3 | tgw3 | qgl3 | gw2 | tgw6 | qlgy3 | GW8 | gw5 | ||
| IR6 | 23.23±0.29 | 4.06±0.02 | 9.20±0.04 | 2.27±0.01 | gs3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | GW5/gw5 | ||
| 华占 | 19.03±0.38 | 3.88±0.02 | 8.29±0.03 | 2.15±0.01 | gs3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | GW5 | ||
| 中花11 | 23.94±0.21 | 2.19±0.01 | 6.92±0.01 | 3.17±0.02 | GS3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | gw8 | gw5 | ||
| 02428 | 23.86±0.20 | 2.32±0.02 | 6.87±0.01 | 2.98±0.02 | GS3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | gw8 | gw5 | ||
| NJ11 | 25.34±0.13 | 2.59±0.01 | 7.49±0.01 | 2.90±0.01 | GS3 | TGW3 | qGL3 | GW2 | tgw6 | qLGY3 | GW8 | gw5 | ||
| 玉针香 | 27.17±0.07 | 5.63±0.02 | 11.65±0.04 | 2.08±0.01 | gs3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | GW5/gw5 | ||
| IR24 | 24.49±0.09 | 3.63±0.02 | 8.81±0.04 | 2.44±0.02 | gs3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | GW5/gw5 | ||
| IR26 | 23.93±0.10 | 3.6±0.01 | 8.76±0.03 | 2.44±0.01 | gs3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | GW5/gw5 | ||
| IR64 | 24.57±0.12 | 4.01±0.02 | 9.07±0.03 | 2.27±0.01 | gs3 | TGW3 | qGL3 | GW2 | TGW6 | qLGY3 | GW8 | GW5/gw5 | ||
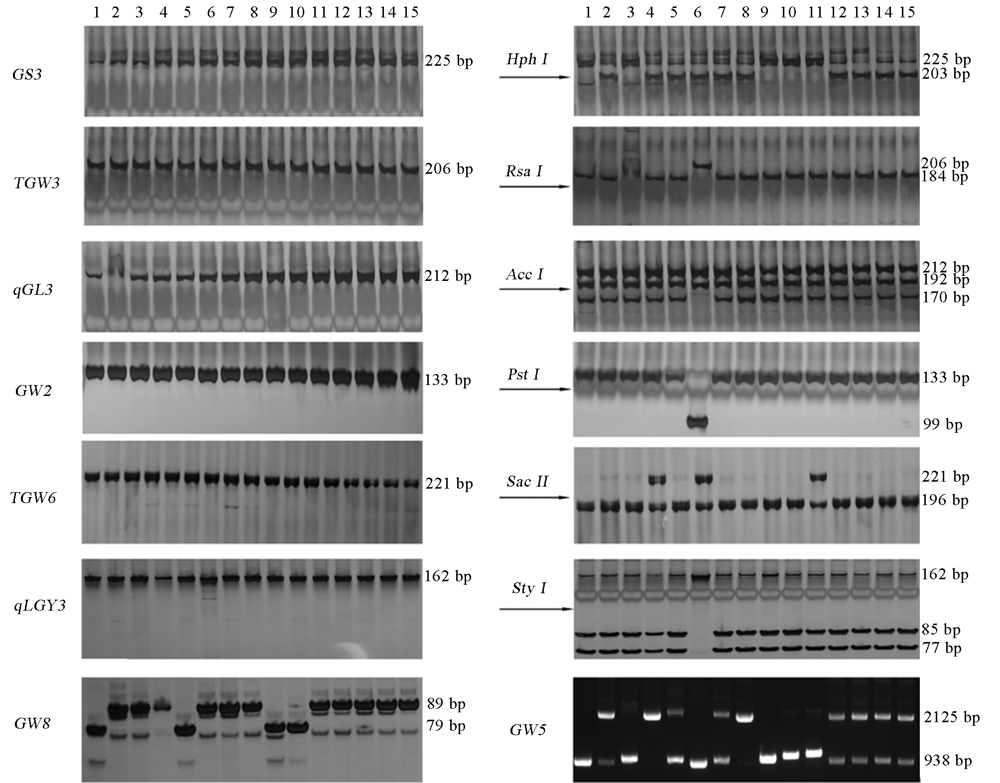
Fig. 5. Typing of eight grain shape genes detected by dCAPS functional marker. 1-15 are the germplasm number in this test, as follows: 1, Nipponbare; 2, 9311; 3, Zhonghui 9308; 4, Zhonghui 8015; 5, Changlijing; 6, BG1; 7, IR56; 8, Huazhan; 9, Zhonghua 11; 10, 02428; 11, Nanjing 11; 12, Yuzhenxiang; 13, IR24; 14, IR26; 15, IR64.
| [1] | Li N, Xu R, Duan P G, Li Y H.Control of grain size in rice[J]. Plant Reproduction, 2018, 31(3): 237-251. |
| [2] | Mao H L, Sun S Y, Yao J L, Yu S B, Xu C G, Li X H, Zhang Q F.Linking differential domain functions of the GS3 protein to natural variation of grain size in rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(45): 19579-19584. |
| [3] | Fan C C, Xing Y Z, Mao H L, Lu T T, Han B, Xu C G, Li X H, Zhang Q F.GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein[J]. Theoretical and Applied Genetics, 2006, 112(6): 1164-1171. |
| [4] | Fan C C, Yu S B, Wang C R, Xing Y Z.A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker[J]. Theoretical and Applied Genetics, 2009, 118(3): 465-472. |
| [5] | Takano-Kai N, Jiang H, Kubo T, Sweeney M, Matsumoto T, Kanamori H, Padhukasahasram B, Bustaamante C, Yoshimura A, Kazuyuki. Evolutionary history of GS3, a gene conferring grain length in rice[J]. Genetics, 2009, 182(4):1323-1334. |
| [6] | Wang C R, Chen S, Yu S B.Functional markers developed from multiple loci in GS3 for fine marker-assisted selection of grain length in rice[J]. Theoretical and Applied Genetics, 2011, 122(5): 905-913. |
| [7] | Qi P, Lin Y S, Song X J, Shen J B, Huang W, Shan J X, Zhu M Z, Jiang L W, Gao J P, Lin H X.The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3[J]. Cell Research, 2012, 22(12): 1666-1680. |
| [8] | Gao X Y, Zhang J Q, Zhang X J, Zhou J, Jiang Z S, Huang P, Tang Z B, Bao Y M, Cheng J P, Tang H J, Zhang W H, Zhang H H, Huang J.Rice qGL3/OsPPKL1 functions with the GSK3/SHAGGY-like kinase OsGSK3 to modulate brassinosteroid signaling[J]. Plant Cell, 2019, 31(5): 1077-1093. |
| [9] | Zhang X J, Wang J F, Huang J, Lan H X, Wang C L, Yin C F, Wu Y Y, Tang H J, Qian Q, Li J Y, Zhang H S.Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(52): 21534-21539. |
| [10] | Hu Z J, He H H, Zhang S Y, Sun F, Xin X Y, Wang W X, Qian X, Yang J S, Luo X J.A Kelch motif-containing serine/threonine protein phosphatase determines the large grain QTL trait in rice.Journal of Integrative Plant Biology, 2012, 54(12): 979-990. |
| [11] | Liu Q, Han R X, Wu K, Zhang J Q, Ye Y F, Wang S S, Chen J F, Pan Y J, Li Q, Xu X P, Zhou J W, Tao D Y, Wu Y J, Fu X D.G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice[J]. Nature Communications, 2018, 9(1): 852. |
| [12] | Wang C S, Tang S C, Zhan Q L, Hou Q Q, Zhao Y, Zhao Q, Feng Q, Zhou C C, Lyu D F, Cui L L, Li Y, Miao J S, Zhu C R, Lu Y Q, Wang Y C, Wang Z Q, Zhu J J, Shangguan Y Y, Gong J Y, Yang S H, Wang W Q, Zhang J F, Xie H A, Huang X H, Han B.Dissecting a heterotic gene through GradedPool-Seq mapping informs a rice-improvement strategy[J]. Nature Communications, 2019, 10(1): 2982. |
| [13] | Song X J, Huang W, Shi M, Zhu M Z, Lin H X.A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase.Nature Genetics, 2007, 39(5): 623-630. |
| [14] | Liu J F, Chen J, Zheng X M, Wu F Q, Lin Q B, Heng Y Q, Tian P, Cheng Z J, Yu X W, Zhou K N, Zhang X, Guo X P, Wang J L, Wang H Y, Wan J M.GW5 acts in the brassinosteroid signaling pathway to regulate grain width and weight in rice[J]. Nature Plants, 2017, 10(4): 17043. |
| [15] | Duan P G, Xu J G, Zeng D L, Zhang B L, Geng M F, Zhang G Z, Huang K, Huang L J, Xu R, Ge S, Qian Q, Li Y H.Natural variation in the promoter of GSE5 contributes to grain size diversity in rice[J]. Molecular Plant, 2017, 10(5): 685-694. |
| [16] | Wang S K, Wu K, Yuan Q B, Liu X Y, Liu Z B, Lin X Y, Zeng R Z, Zhu H T, Dong G J, Qian Q, Zhang G Q, Fu X D.Control of grain size, shape and quality by OsSPL16 in rice[J]. Nature Genetics, 2012, 44(8): 950-954. |
| [17] | Zhao D S, Li Q F, Zhang C Q, Zhang C, Yang Q Q, Pan L X, Ren X Y, Lu J, Gu M H, Liu Q Q.GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nature Communications, 2018, 9(1): 1240. |
| [18] | Hu Z J, Lu S J, Wang M J, He H H, Sun L, Wang H R, Liu X H, Jiang L, Sun J L, Xin X Y, Kong W, Chu C C, Xue H W, Yang J S, Luo X J, Liu J X.A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice[J]. Molecular Plant, 2018, 11(5): 736-749. |
| [19] | Ying J Z, Ma M, Bai C, Huang X H, Liu J L, Fan Y Y, Song X J.TGW3, a major QTL that negatively modulates grain length and weight in rice[J]. Molecular Plant, 2018, 11(5): 750-753. |
| [20] | Xia D, Zhou H, Liu R J, Dan W H, Li P B, Wu B, Chen J X, Wang L Q, Gao G J, Zhang Q L, He Y Q.GL3.3, a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically interacts with GS3 to produce extra-long grains in rice[J]. Molecular Plant, 2018, 11(5): 754-756. |
| [21] | Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B I, Onishi A, Miyagawa H, Katoh E.Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield[J]. Nature Genetics, 2013, 45(6): 707-711. |
| [22] | Gao X Y, Zhang X J, Lan H X, Huang J, Wang J F, Zhang H S.The additive effects of GS3 and qGL3 on rice grain length regulation revealed by genetic and transcriptome comparisons[J]. BMC Plant Biology, 2015, 15:156. |
| [23] | Wang S K, Li S, Liu Q, Wu K, Zhang J Q, Wang S S, Wang Y, Chen X B, Zhang Y, Gao C X, Wang F, Huang H X, Fu X D.The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality[J]. Nature Genetics, 2015, 47(8): 949-954. |
| [24] | 陈杰, 朱业宝, 孙新立, 翁群清, 张国杰, 梁康迳. 一个特大粒水稻种质粒形相关基因的检测[J]. 分子植物育种, 2017, 15(2): 576-581. |
| Chen J, Zhu Y B, Sun X L, Weng Q Q, Zhang G J, Liang K J.Detection of grain shape related genes in a large grain rice germplasm[J]. Molecular Plant Breeding, 2017, 15(2): 576-581. | |
| [25] | Rogers S O, Bendich A J.Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues[J]. Plant Molecular Biology, 1985, 5(2): 69-76. |
| [26] | Neff M M, Turk E, Kalishman M.Web-based primer design for single nucleotide polymorphism analysis[J]. Trends in Genetics, 2002, 18(12): 613-615. |
| [27] | Zhang L, Ma B, Bian Z, Li X Y, Zhang C Q, Liu J Y, Li Q, Liu Q Q, He Z H.Grain size selection using novel functional markers targeting 14 genes in rice[J]. Rice, 2020, 13(1): 63. |
| [28] | 肖国樱, 肖友伦, 李锦江, 邓力华, 翁绿水, 孟秋成, 于江辉. 高效是当前水稻育种的主导目标[J].中国水稻科学, 2019, 33(4): 287-292. |
| Xiao G Y, Xiao Y L, Li J J, Deng L H, Weng L S, Meng Q C, Yu J H.High efficiency is the leading goal of rice breeding at present[J]. Chinese Journal of Rice Science, 2019, 33(4): 287-292. (in Chinese with English abstract) | |
| [29] | 裔传灯, 王德荣, 蒋伟, 李玮, 成晓俊, 王颖, 周勇, 梁国华, 顾铭洪. 水稻粒形基因GW8的功能标记开发和单体型鉴定[J]. 作物学报, 2016, 42(9): 1291-1297. |
| Yi C D, Wang D R, Jiang W, Li W, Cheng X J, Wang Y, Zhou Y, Liang G H, Gu M H.Development of functional markers and haplotype identification of rice grain shape gene GW8[J]. Acta Agronomica Sinica, 2016, 42(9): 1291-1297. (in Chinese with English abstract) | |
| [30] | 刘李鑫哲, 何宇涵, 炎会敏, 李俊周, 赵全志. 水稻粒宽基因GW5和GW8功能标记的开发与多重PCR检测[J]. 分子植物育种, 2019, 17(13):4280-4288. |
| LiuLi X Z, He Y H, Yan H M, Li J Z, Zhao Q Z. Development of functional markers of rice grain width genes GW5 and GW8 and detection of multiple PCR[J]. Molecular Plant Breeding, 2019, 17(13): 4280-4288. | |
| [31] | 陈深广, 周屹峰, 赵霏, 金亮, 沈圣泉. 利用Wx和fgr基因双功能性标记高效选育优质水稻保持系[J]. 中国水稻科学, 2011, 25(1): 31-36. |
| Chen S G, Zhou Y F, Zhao F, Jin L, Shen S Q.High quality rice maintainer lines were selected by using bifunctional markers of Wx and fgr genes[J]. Chinese Journal of Rice Science, 2011, 25(1): 31-36. (in Chinese with English abstract) | |
| [32] | 李扬, 徐小艳, 严明, 冯芳君, 马孝松, 梅捍卫. 利用GS3基因功能性分子标记改良水稻粒型的研究[J]. 上海农业学报, 2016, 32(1): 1-5. |
| Li Y, Xu X Y, Yan M, Feng F J, Ma X S, Mei H W.Study on the improvement of rice grain shape by functional molecular markers of GS3 gene[J]. Shanghai Journal of Agriculture, 2016, 32(1):1-5. (in Chinese with English abstract) |
| [1] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [2] | WANG Jun, ZHOU Jing, TAO Yajun, LI Wenqi, ZHU Jianping, FAN Fangjun, WANG Fangquan, XU Yang, CHEN Zhihui, JIANG Yanjie, LI Xia, YANG Jie. Development of HRM-based Functional Marker for Gelatinization Temperature Gene ALK in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(1): 106-110. |
| [3] | MA Zhaohui, SHI Yihan, CHENG Haitao, SONG Wenwen, LU Lianji, LIU Renguang, LÜ Wenyan. Effects of Embryo Morphology and Endosperm Composition on Embryo-remaining Characteristics in Rice [J]. Chinese Journal OF Rice Science, 2023, 37(3): 265-275. |
| [4] | WANG Shiguang, LU Zhanhua, LIU Wei, LU Dongbai, WANG Xiaofei, FANG Zhiqiang, WU Haoxiang, HE Xiuying. Generating Guangdong Simiao Rice Germplasms by Applying CRISPR/Cas9 Gene Editing and Marker-assisted Selection Technology [J]. Chinese Journal OF Rice Science, 2023, 37(1): 29-36. |
| [5] | XU Yunji, TANG Shupeng, JIAN Chaoqun, CAI Wenlu, ZHANG Weiyang, WANG Zhiqin, YANG Jianchang. Roles of Polyamines and Ethylene in Grain Filling, Grain Weight and Quality of Rice [J]. Chinese Journal OF Rice Science, 2022, 36(4): 327-335. |
| [6] | HUANG Tao, WANG Yanning, ZHONG Qi, CHENG Qin, YANG Mengmeng, WANG Peng, WU Guangliang, HUANG Shiying, LI Caijing, YU Jianfeng, HE Haohua, BIAN Jianmin. Mapping and Analysis of QTLs for Rice Grain Weight and Grain Shape Using Chromosome Segment Substitution Line Population [J]. Chinese Journal OF Rice Science, 2022, 36(2): 159-170. |
| [7] | Haoliang YAN, Song WANG, Xueyan WANG, Chengcheng DANG, Meng ZHOU, Rongrong HAO, Xiaohai TIAN. Performance of Different Rice Varieties Under High Temperature and Its Relationship with Field Meteorological Factors [J]. Chinese Journal OF Rice Science, 2021, 35(6): 617-628. |
| [8] | Yudong CAO, Xiangyi XIAO, Naizhong YE, Xiaowen DING, Xiaoxuan YI, Jinling LIU, Yinghui XIAO. Auxin Regulator OsGRF4 Simultaneously Regulates Rice Grain Shape and Blast Resistance [J]. Chinese Journal OF Rice Science, 2021, 35(6): 629-638. |
| [9] | Chengxing DU, Huali ZHANG, Dongqing DAI, Mingyue WU, Minmin LIANG, Junyu CHEN, Liangyong MA. QTL Analysis for Grain Weight and Shape and Validation of qTGW1.2/qGL1.2 [J]. Chinese Journal OF Rice Science, 2021, 35(4): 359-372. |
| [10] | Xuemei DENG, Peng HU, Yueying WANG, Yi WEN, Yiqing TAN, Hao WU, Kaixiong WU, Junge WANG, Linlin HOU, Lixin ZHU, Li ZHU, Guang CHEN, Dali ZENG, Guangheng ZHANG, Longbiao GUO, Zhenyu GAO, Deyong REN, Qian QIAN, Jiang HU. Identification and Fine Mapping of a Grain Width Mutant gw4 in Rice [J]. Chinese Journal OF Rice Science, 2021, 35(3): 259-268. |
| [11] | Shanbin XU, Hongliang ZHENG, Lifeng LIU, Qingyun BU, Xiufeng Li, Detang ZOU. Improvement of Grain Shape and Fragrance by Using CRISPR/Cas9 System [J]. Chinese Journal OF Rice Science, 2020, 34(5): 406-412. |
| [12] | Guangda WANG, Peng GAO, Wenyan YANG, Ao CUI, Jianhua ZHAO, Zhiming FENG, Wenlei CAO, Zongxiang CHEN, Shimin ZUO. Development and Utilization of Functional Markers for Imidazolinone Herbicides Resistance Gene in japonica Rice Variety Jinjing 818 [J]. Chinese Journal OF Rice Science, 2020, 34(4): 316-324. |
| [13] | Panpan LI, Yujun ZHU, Liang GUO, Jieyun ZHUANG, Yeyang FAN. Fine Mapping of qGL1.1, a Minor QTL for Grain Length, Using Near Isogenic Lines Derived from Residual Heterozygotes in Rice [J]. Chinese Journal OF Rice Science, 2020, 34(2): 125-134. |
| [14] | Andong ZHU, Zhichao SUN, Yujun ZHU, Hui ZHANG, Xiaojun NIU, Yeyang FAN, Zhenhua ZHANG, Jieyun ZHUANG. Identification of QTL for Grain Weight and Grain Shape Using Populations Derived from Residual Heterozygous Lines of indica Rice [J]. Chinese Journal OF Rice Science, 2019, 33(2): 144-151. |
| [15] | Xi LIU, Changling MOU, Chunlei ZHOU, Zhijun CHENG, Ling JIANG, Jianmin WAN. Research Progress on Cloning and Regulation Mechanism of Rice Grain Shape Genes [J]. Chinese Journal OF Rice Science, 2018, 32(1): 1-11. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||