
Chinese Journal OF Rice Science ›› 2021, Vol. 35 ›› Issue (6): 535-542.DOI: 10.16819/j.1001-7216.2021. 210205
• 研究报告 • Previous Articles Next Articles
Xianmei WU1,#, Sanfeng LI1,#, Ping HU1, Rui HE1, Ran JIAO2, Yijian MAO1, Caolin LU1, Juan HU2, Han LIN2, Rongliang WU1, Xudong ZHU1, Yuchun RAO2,*( ), Yuexing WANG1,*(
), Yuexing WANG1,*( )
)
Received:2021-02-16
Revised:2021-03-23
Online:2021-11-10
Published:2021-11-10
Contact:
Yuchun RAO, Yuexing WANG
About author:#These authors contributed equally to this work
吴先美1,#, 李三峰1,#, 胡萍1, 何瑞1, 焦然2, 毛一剑1, 鲁草林1, 胡娟2, 林晗2, 吴荣梁1, 朱旭东1, 饶玉春2,*( ), 王跃星1,*(
), 王跃星1,*( )
)
通讯作者:
饶玉春,王跃星
作者简介:#共同第一作者
基金资助:Xianmei WU, Sanfeng LI, Ping HU, Rui HE, Ran JIAO, Yijian MAO, Caolin LU, Juan HU, Han LIN, Rongliang WU, Xudong ZHU, Yuchun RAO, Yuexing WANG. Cloning and Functional Analysis of Rice Tillering Regulatory Gene HTD3[J]. Chinese Journal OF Rice Science, 2021, 35(6): 535-542.
吴先美, 李三峰, 胡萍, 何瑞, 焦然, 毛一剑, 鲁草林, 胡娟, 林晗, 吴荣梁, 朱旭东, 饶玉春, 王跃星. 水稻分蘖调控基因HTD3的克隆与功能研究[J]. 中国水稻科学, 2021, 35(6): 535-542.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2021. 210205
| 标记 Marker | 正向引物 Forward primer (5′→3′) | 反向引物 Reverse primer (5′→3′) |
|---|---|---|
| B12-6 | TGAAGCGGTTTGACTTTGACC | GGGGTGAAAACTGGTAGGGT |
| B12-9 | TCCTTCTCGTTTATGAACTTATGG | TAGAGCAAAGCAGAGCCCAG |
| M9 | TAATCCCCTGCACTCCATCC | GTGAACAACCAGCCGAGAAT |
| M17 | GACTTTCTAGCATTGCCCACA | GCATTAACTGGGGCCATTGT |
| M25 | GAGACGGCCAGCTTAGGTAG | CAGTGCTACAGAAACAGGGC |
| M29 | CCGAACTCCAGTTTGTGAGG | CGGAAATCTGACGCTGGTAT |
| M35 | AAAAGAACAACACAGCCCCT | ACAAGGAGCAATCTGGACCA |
| CM5 | TTGTGAACAAGAGCCAACGG | CGCTGTTGGGCATTCTTTAAAG |
| CM8 | GTGTGATCCATGGGTAGCCT | CAGAGCCCTATTAGTCTATTGCT |
| CM10 | TCACATGATACCTCGCGAGT | CAGACGATTCTACACAACAGGA |
| CM13 | AGCAGCCAAGATTAAGGAGGA | CGTCAGAGTGATTAGCAAAAGGA |
Table 1 Sequences of markers for HTD3 mapping.
| 标记 Marker | 正向引物 Forward primer (5′→3′) | 反向引物 Reverse primer (5′→3′) |
|---|---|---|
| B12-6 | TGAAGCGGTTTGACTTTGACC | GGGGTGAAAACTGGTAGGGT |
| B12-9 | TCCTTCTCGTTTATGAACTTATGG | TAGAGCAAAGCAGAGCCCAG |
| M9 | TAATCCCCTGCACTCCATCC | GTGAACAACCAGCCGAGAAT |
| M17 | GACTTTCTAGCATTGCCCACA | GCATTAACTGGGGCCATTGT |
| M25 | GAGACGGCCAGCTTAGGTAG | CAGTGCTACAGAAACAGGGC |
| M29 | CCGAACTCCAGTTTGTGAGG | CGGAAATCTGACGCTGGTAT |
| M35 | AAAAGAACAACACAGCCCCT | ACAAGGAGCAATCTGGACCA |
| CM5 | TTGTGAACAAGAGCCAACGG | CGCTGTTGGGCATTCTTTAAAG |
| CM8 | GTGTGATCCATGGGTAGCCT | CAGAGCCCTATTAGTCTATTGCT |
| CM10 | TCACATGATACCTCGCGAGT | CAGACGATTCTACACAACAGGA |
| CM13 | AGCAGCCAAGATTAAGGAGGA | CGTCAGAGTGATTAGCAAAAGGA |
| 引物名称 Primer name | 引物序列 Primer sequences (5′→3′) |
|---|---|
| HTD3-CPT-EcoRⅠ-F | ACGAATTCGAGCTCGGTACCTGGTTCTGTGACTAAAGCGC |
| HTD3-CPT-HindⅢ-R | GGCCAGTGCCAAGCTTTCTCCGGGGCCCTGAATATTCCTCT |
Table 2 Primers for amplifying the complete expression unit of HTD3.
| 引物名称 Primer name | 引物序列 Primer sequences (5′→3′) |
|---|---|
| HTD3-CPT-EcoRⅠ-F | ACGAATTCGAGCTCGGTACCTGGTTCTGTGACTAAAGCGC |
| HTD3-CPT-HindⅢ-R | GGCCAGTGCCAAGCTTTCTCCGGGGCCCTGAATATTCCTCT |
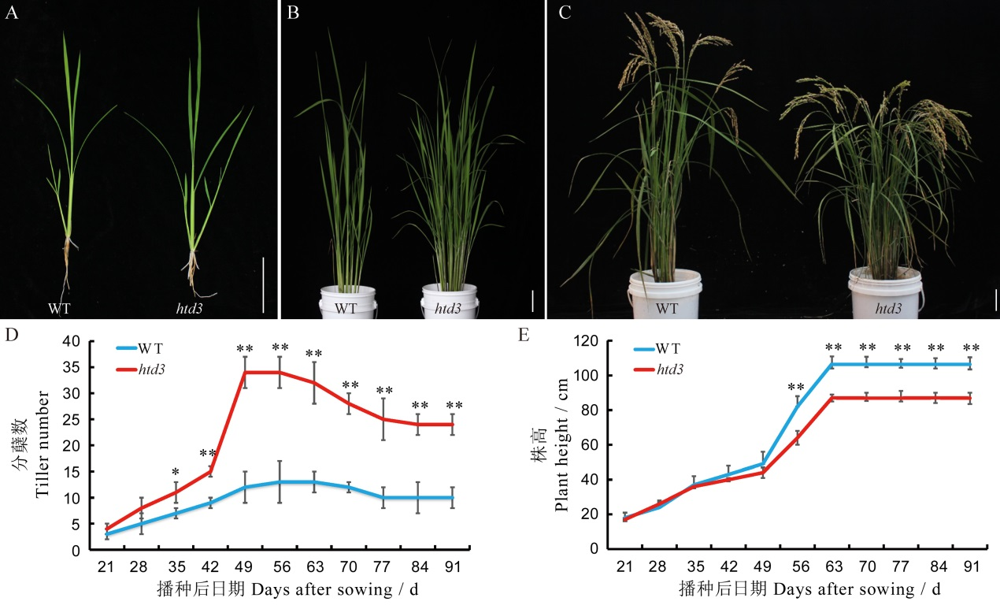
Fig. 1. Morphological characterization of WT and htd3. A~C, Plant phenotypes of wild-type (WT) and htd3 in the seedling stage (A), tillering stage (B) , and mature stage (C) , bar= 6 cm; D~E, Tiller number (D) and plant height (E) of WT and htd3 after sowing. Values are means ± SD (n=15), t-test, *, ** indicate significant difference at P<0.05 and P<0.01, respectively.
| 农艺性状Agronomic trait | 野生型WT | 突变体htd3 |
|---|---|---|
| 株高Plant height /cm | 97.08±2.78 | 83.76±1.54* |
| 有效穗数Effective panicles | 11.0±1.0 | 28.0±2.0** |
| 穗长Panicle length/cm | 21.52±0.98 | 20.82±0.62 |
| 一次枝梗数 Primary branches | 12.0±2.0 | 9.0±1.0* |
| 二次枝梗数Secondary branches | 26.0±3.0 | 23.0±4.0 |
| 每穗粒数Grains per panicle | 121.0±11.0 | 104.0±16.0* |
| 每株总粒数Total grains per plant | 1331±28 | 2018±36** |
| 千粒重1000-grain weight/g | 23.7±2.1 | 23.9±1.3 |
| 结实率Seed setting rate/% | 87.2±5.8 | 90.0±2.4 |
Table 3 Comparison of agronomic traits between WT and htd3.
| 农艺性状Agronomic trait | 野生型WT | 突变体htd3 |
|---|---|---|
| 株高Plant height /cm | 97.08±2.78 | 83.76±1.54* |
| 有效穗数Effective panicles | 11.0±1.0 | 28.0±2.0** |
| 穗长Panicle length/cm | 21.52±0.98 | 20.82±0.62 |
| 一次枝梗数 Primary branches | 12.0±2.0 | 9.0±1.0* |
| 二次枝梗数Secondary branches | 26.0±3.0 | 23.0±4.0 |
| 每穗粒数Grains per panicle | 121.0±11.0 | 104.0±16.0* |
| 每株总粒数Total grains per plant | 1331±28 | 2018±36** |
| 千粒重1000-grain weight/g | 23.7±2.1 | 23.9±1.3 |
| 结实率Seed setting rate/% | 87.2±5.8 | 90.0±2.4 |
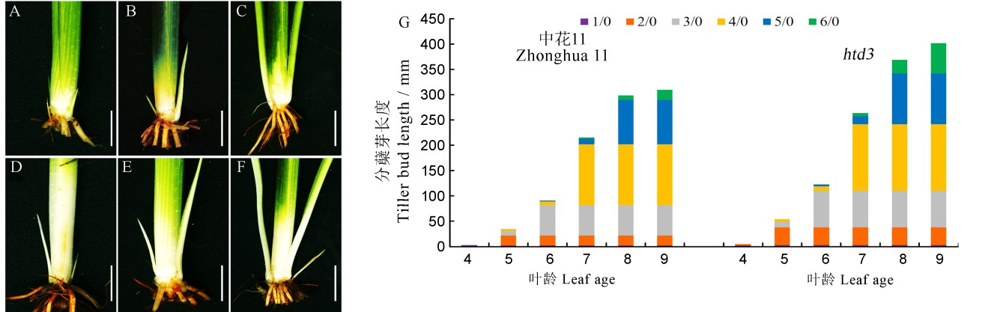
Fig. 2. Phenotypic observation of tillering buds of WT and htd3. A~F, Phenotype of tillering buds of WT and htd3 at five-, six- and seven-leaf stage; A-C represents WT; D-F represents htd3; bars=1 cm; G, Tillering bud length of WT and htd3 at each leaf stage, in n/0, n represents the leaf position, 0 represents the main stem, 1/0 represents the tiller bud corresponding to the first leaf of the main stem, 2/0 represents the tiller bud corresponding to the second leaf of the main stem, and so on.
| 组合Cross | F1表型 Phenotype of F1 plants | F2群体 F2 population | χ2 (3:1) | χ20. 05 | ||
|---|---|---|---|---|---|---|
| 总株数 No. of plants | 正常分蘖株数 No. of normal plants | 多分蘖株数 No. of high-tillering plants | ||||
| 中花11/ TN1 Zhonghua 11/TN1 | 正常分蘖Normal tiller | 864 | 666 | 198 | 2 | 3. 84 |
Table 4 Genetic analysis of rice high-tillering mutant htd3.
| 组合Cross | F1表型 Phenotype of F1 plants | F2群体 F2 population | χ2 (3:1) | χ20. 05 | ||
|---|---|---|---|---|---|---|
| 总株数 No. of plants | 正常分蘖株数 No. of normal plants | 多分蘖株数 No. of high-tillering plants | ||||
| 中花11/ TN1 Zhonghua 11/TN1 | 正常分蘖Normal tiller | 864 | 666 | 198 | 2 | 3. 84 |
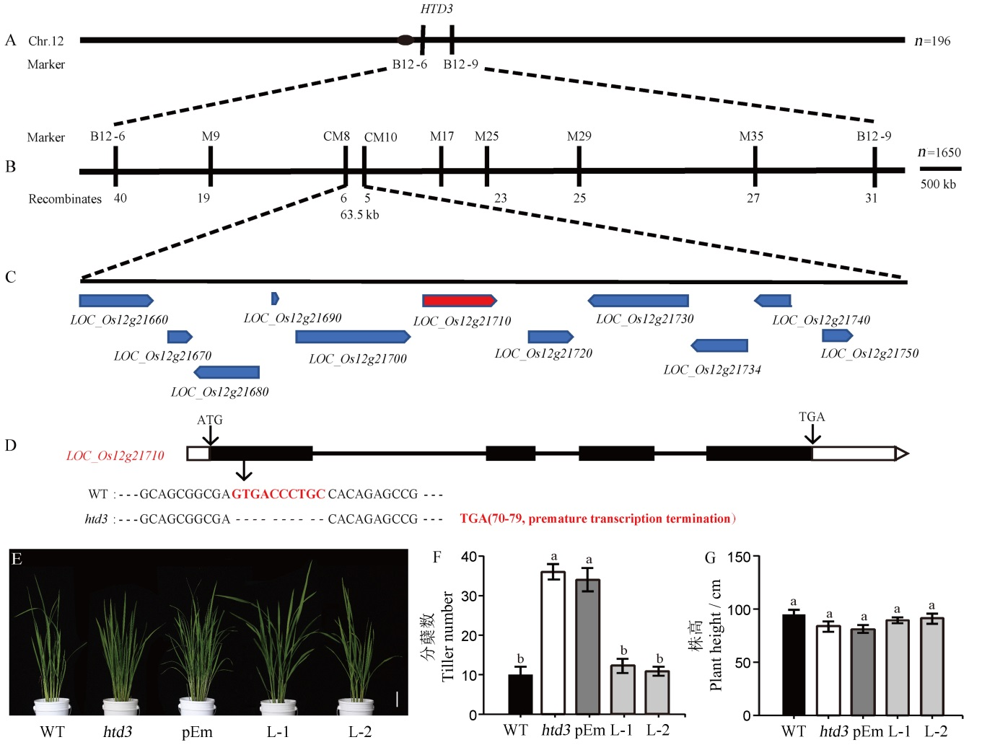
Fig. 3. Map-based cloning of HTD3 and verification of complementary transgene. A, Initial location of HTD3; B, Fine mapping of HTD3; C, Predicted ORFs in the location interval; D, Gene structure of the candidate gene LOC_Os12g21710 and the sequence difference between WT and htd3, in which black boxes, white boxes, black lines represent exons, UTR and introns, respectively; E, Phenotype of WT, htd3 and complementary transgenic T2 generation plants in the tilling stage, bar=6 cm; F~G, Number of tillers (F) and plant height (G) of WT, htd3 and complementary transgenic T2 generation plants in the tillering stage, the data are means ± standard deviation (n=5). Significant difference by Duncan’s multiple range test. The same letters indicate no significant difference at P<0.05.
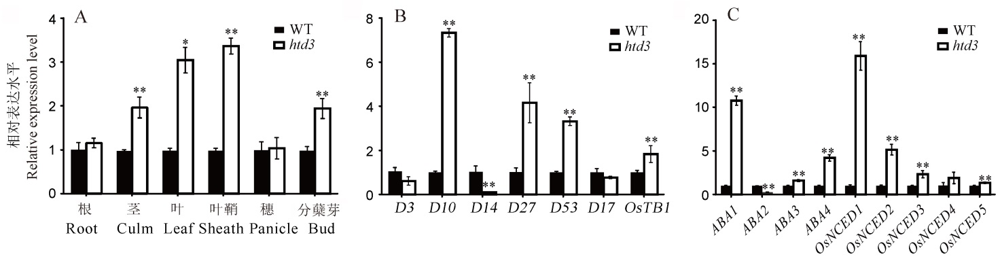
Fig. 4. Expression analysis of HTD3 and hormone related genes. A, HTD3 relative expression level in various tissues of WT and htd3; B-C, Relative expression of SLs (B) and ABA (C) related genes in WT and htd3. RNA was isolated from WT and htd3 leaves in B and C. Expression levels are represented as relative to the corresponding genes in WT (set as reference value of 1), and data are shown as means ± SD from three biological replicates. Asterisks indicate statistical significance between the WT and the mutant, as determined by Student’s t-test (*P < 0.05; **P < 0.01).
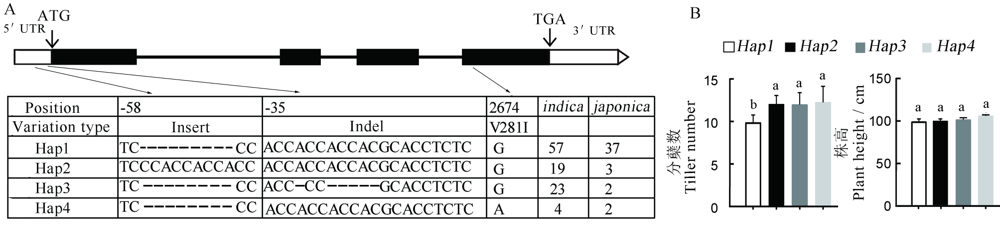
Fig. 5. Analysis of the HTD3 gene haplotypes. A, Gene structure of the HTD3, and nucleotide polymorphisms in the HTD3 5'-UTR and coding region, exons are indicated by black boxes, UTRs are indicated by white boxes, and the black lines between black boxes represent introns, hap means haplotypes, the number represents the base position relative to the start codon ATG. B, Tiller number and plant height of each haplotype. Significant difference by Duncan’s multiple range test. The same letters indicate no significant difference at P<0.05.
| [1] | 李学勇, 钱前, 李家洋. 水稻分蘖的分子机理研究[J]. 中国科学院院刊, 2003, 18(4): 274-276. |
| Li X Y, Qian Q, Li J Y.Progress in elucidating the molecular mechanism of rice tillering[J]. Bulletin of the Chinese Academy of Sciences, 2003, 18(4): 274-276. (in Chinese with English abstract) | |
| [2] | Wu T, Shen Y, Zheng M, Yang C, Chen Y, Feng Z, Liu X, Liu S, Chen Z, Lei C, Wang J, Jiang L, Wan J.Gene SGL, encoding a kinesin-like protein with transactivation activity, is involved in grain length and plant height in rice[J]. Plant Cell Reports, 2014, 33(2): 235-244. |
| [3] | Li X Y, Qian Q, Fu Z M, Wang Y H, Xiong G S, Zeng D L, Wang X Q, Liu X F, Teng S, Hiroshi F, Yuan M, Luo D, Han B, Li J Y.Control of tillering in rice[J]. Nature, 2003, 422(6932): 618-621. |
| [4] | Xu C, Wang Y, Yu Y, Duan J, Liao Z, Xiong G, Meng X, Liu G, Qian Q, Li J.Degradation of MONOCULM 1 by APC/C(TAD1) regulates rice tillering[J]. Nature Communications, 2012, 3(1): 1-9. |
| [5] | Liang W H, Shang F, Lin Q T, Lou C, Zhang J.Tillering and panicle branching genes in rice[J]. Gene, 2014, 537(1): 1-5. |
| [6] | Kim H, Hwang H, Hong J W, Lee Y N, Ahn I P, Yoon I S, Yoo S D, Lee S, Lee S C, Kim B G.A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth[J]. Journal of Experimental Botany, 2012, 63(2): 1013-1024. |
| [7] | Ljung K, Bhalerao R P, Sandberg G.Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J, 2001, 28(4):465-474. |
| [8] | Lee M, Jung J H, Han D Y, Seo P J, Park W J, Park C M.Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis[J]. Planta, 2012, 235(5): 923-938. |
| [9] | Xu M, Zhu L, Shou H, Wu P.A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice[J]. Plant Cell Physiology, 2005, 46(10): 1674-1681. |
| [10] | Xu J X, Ding C Q, Ding Y F, Tang S, Zha M R, Luo B J, Wang S H.A proteomic approach to analyze differential regulation of proteins during bud outgrowth under apical dominance based on the auxin transport canalization model in rice (Oryza sativa L.)[J]. Journal of Plant Growth Regulation, 2015, 34(1): 122-136. |
| [11] | Lin H, Wang R, Qian Q, Yan M, Meng X, Fu Z, Yan C, Jiang B, Su Z, Li J, Wang Y.DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth[J]. Plant Cell, 2009, 21(5): 1512-1525. |
| [12] | Zou J, Zhang S, Zhang W, Li G, Chen Z, Zhai W, Zhao X, Pan X, Xie Q, Zhu L.The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds[J]. Plant Journal, 2006, 48(5): 687-698. |
| [13] | Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J.DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice[J]. Plant Journal, 2007, 51(6): 1019-1029. |
| [14] | Fang Z, Ji Y, Hu J, Guo R, Sun S, Wang X.Strigolactones and brassinosteroids antagonistically regulate the stability of the D53-OsBZR1 complex codetermine FC1 expression in rice tillering[J]. Molecular Plant, 2020, 13(4): 586-597. |
| [15] | Zhao J, Wang T, Wang M, Zhao J, Wang T, Wang M, Liu Y, Yuan S, Gao Y, Yin L, Sun W, Wan J, Li X.DWARF3 participates in an SCF complex and associates with DWARF14 to suppress rice shoot branching[J]. Plant Cell Physiology, 2014, 55(6): 1096-1109. |
| [16] | Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J.d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers[J]. Plant Cell Physiology, 2009, 50(8): 1416-1424. |
| [17] | Liu W, Wu C, Fu Y, Hu G, Si H, Zhu L, Luan W, He Z, Sun Z.Identification and characterization of HTD2: A novel gene negatively regulating tiller bud outgrowth in rice[J]. Planta, 2009, 230(4): 649-658. |
| [18] | Gao Z, Qian Q, Liu X, Yan M, Feng Q, Dong G, Liu J, Han B.Dwarf 88, a novel putative esterase gene affecting architecture of rice plant[J]. Plant Molecular Biology, 2009, 71(3): 265-276. |
| [19] | Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J.Suppression of tiller bud activity in tillering dwarf mutants of rice[J]. Plant Cell Physiology, 2005, 46(1): 79-86. |
| [20] | De S A, Clavé G, Badet-Denisot M A, Pillot J P, Cornu D, Le Caer J P, Burger M, Pelissier F, Retailleau P, Turnbull C, Bonhomme S, Chory J, Rameau C, Boyer F D. An histidine covalent receptor and butenolide complex mediates strigolactone perception[J]. Nature Chemical Biology, 2016, 12(10): 787-794. |
| [21] | Sharma R, De V D, Sharma M K, Ronald P C.Recent advances in dissecting stress-regulatory crosstalk in rice[J]. Molecular Plant, 2013, 6(2): 250-260. |
| [22] | Yao R, Ming Z, Yan L, Li S, Wang F, Ma S, Yu C, Yang M, Chen L, Li Y, Yan C, Miao D, Sun Z, Yan J, Sun Y, Wang L, Chu J, Fan S, He W, Deng H, Nan F, Li J, Rao Z, Lou Z, Xie D.DWARF14 is a non-canonical hormone receptor for strigolactone[J]. Nature, 2016, 536(7617): 469-473. |
| [23] | Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N, Wu F, Mao H, Dong W, Gan L, Ma W.D14-SCF(D3)- dependent degradation of D53 regulates strigolactone signaling[J]. Nature, 2013, 504(7480): 406-410. |
| [24] | Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi- Tanaka M, Ashikari M, Matsuoka M, Ueguchi C.The OsTB1 gene negatively regulates lateral branching in rice[J]. Plant Journal, 2003, 33(3): 513-520. |
| [25] | Umehara M, Hanada A, Magome H, Takeda K N, Yamaguchi S.Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice[J]. Plant Cell Physiology, 2010, 51(7): 1118-1126. |
| [26] | Wang Y, Shang L, Yu H, Zeng L, Hu J, Ni S, Rao Y, Li S, Chu J, Meng X, Wang L, Hu P, Yan J, Kang S, Qu M, Lin H, Wang T, Wang Q, Hu X, Chen H, Wang B, Gao Z, Guo L, Xiong G, Li J, Qian Q.A strigolactone biosynthesis gene contributed to the green revolution in rice[J]. Molecular Plant, 2020, 13(6): 923-932. |
| [27] | Liu X, Hu Q, Yan J, Sun K, Liang Y, Jia M, Meng X, Fang S, Wang Y, Jing Y, Liu G, Wu D, Chu C, Smith S M, Chu J, Wang Y, Li J, Wang B.ζ-carotene isomerase suppresses tillering in rice through the coordinated biosynthesis of strigolactone and abscisic acid[J]. Molecular Plant, 2020, 13(12): 1784-1801. |
| [28] | Liu L, Ren M, Peng P, Chun Y, Li L, Zhao J, Fang J, Peng L, Yan J, Chu J, Wang Y, Yuan S, Li X.MIT1, encoding a 15-cis-ζ-carotene isomerase, regulates tiller number and stature in rice[J]. Journal of Genet Genomics, 2021, 48(1): 88-91. |
| [29] | 陈彩艳, 邹军煌, 张淑英, 朱立煌. 独角金内酯能抑制植物的分枝并介导植物与枞枝真菌及寄生植物间的相互作用[J]. 中国科学: 生命科学, 2009(6): 525-533. |
| Chen C, Zou J, Zhang S, Zhu L.Strigolactones are a new-defined class of plant hormones which inhibit shoot branching and mediate the interaction of plant-AM fungi and plant-parasitic weeds[J]. Science in China: Life Sciences, 2009(6): 525-533. (in Chinese with English abstract) | |
| [30] | Wang P, Gao J, Wan C, Zhang F, Xu Z, Huang X, Sun X, Deng X.Divinyl chlorophyll(ide) a can be converted to monovinyl chlorophyll(ide) a by a divinyl reductase in rice[J]. Plant Physiology, 2010, 153(3): 994-1003. |
| [31] | Cline M G, Oh C.A reappraisal of the role of abscisic acid and its interaction with auxin in apical dominance[J]. Annual Botany, 2006, 98(4): 891-897. |
| [32] | 刘杨. 水稻分蘖芽萌发与休眠相互转换的激素学机制[D]. 南京: 南京农业大学, 2011. |
| Liu Y.The Mechanism of hormonal regulation of the transformation between germination and dormancy of rice tiller buds[D]. Nanjing: Nanjing Agricultural University.(in Chinese with English abstract) | |
| [33] | Zang G, Zou H, Zhang Y.The De-Etiolated 1 homolog of Arabidopsis modulates the ABA signaling pathway and ABA biosynthesis in rice[J]. Plant Physiology, 2016, 171(2): 1259-1276. |
| [34] | Bang S W, Park S H, Jeong J S, Kim Y S, Jung H, Ha S H, Kim J K.Characterization of the stress-inducible OsNCED3 promoter in different transgenic rice organs and over three homozygous generations[J]. Planta, 2013, 237(1): 211-224. |
| [1] |
WANG Yichen, ZHU Benshun, ZHOU Lei, ZHU Jun, YANG Zhongnan.
Sterility Mechanism of Photoperiod/Thermo-sensitive Genic Male Sterile Lines and Development and Prospects of Two-line Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(5): 463-474. |
| [2] |
XU Yongqiang XU Jun, FENG Baohua, XIAO Jingjing, WANG Danying, ZENG Yuxiang, FU Guanfu.
Research Progress of Pollen Tube Growth in Pistil of Rice and Its Response to Abiotic stress [J]. Chinese Journal OF Rice Science, 2024, 38(5): 495-506. |
| [3] |
HE Yong, LIU Yaowei, XIONG Xiang, ZHU Danchen, WANG Aiqun, MA Lana, WANG Tingbao, ZHANG Jian, LI Jianxiong, TIAN Zhihong.
Creation of Rice Grain Size Mutants by Editing OsOFP30 via CRISPR/Cas9 System [J]. Chinese Journal OF Rice Science, 2024, 38(5): 507-515. |
| [4] |
LÜ Yang, LIU Congcong, YANG Longbo, CAO Xinglan, WANG Yueying, TONG Yi, Mohamed Hazman, QIAN Qian, SHANG Lianguang, GUO Longbiao.
Identification of Candidate Genes for Rice Nitrogen Use Efficiency by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(5): 516-524. |
| [5] |
YANG Hao, HUANG Yanyan, WANG Jian, YI Chunlin, SHI Jun, TAN Chutian, REN Wenrui, WANG Wenming.
Development and Application of Specific Molecular Markers for Eight Rice Blast Resistance Genes in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(5): 525-534. |
| [6] |
JIANG Peng, ZHANG Lin, ZHOU Xingbing, GUO Xiaoyi, ZHU Yongchuan, LIU Mao, GUO Chanchun, XIONG Hong, XU Fuxian.
Yield Formation Characteristics of Ratooning Hybrid Rice Under Simplified Cultivation Practices in Winter Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(5): 544-554. |
| [7] |
YANG Mingyu, CHEN Zhicheng, PAN Meiqing, ZHANG Bianhong, PAN Ruixin, YOU Lindong, CHEN Xiaoyan, TANG Lina, HUANG Jinwen.
Effects of Nitrogen Reduction Combined with Biochar Application on Stem and Sheath Assimilate Translocation and Yield Formation in Rice Under Tobacco-rice Rotation [J]. Chinese Journal OF Rice Science, 2024, 38(5): 555-566. |
| [8] |
XIONG Jiahuan, ZHANG Yikai, XIANG Jing, CHEN Huizhe, XU Yicheng, WANG Yaliang, WANG Zhigang, YAO Jian, ZHANG Yuping .
Effect of Biochar-based Fertilizer Application on Rice Yield and Nitrogen Utilization in Film- mulched PaddyFields [J]. Chinese Journal OF Rice Science, 2024, 38(5): 567-576. |
| [9] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [10] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [11] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [12] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [13] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [14] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [15] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||