
Chinese Journal OF Rice Science ›› 2020, Vol. 34 ›› Issue (3): 278-286.DOI: 10.16819/j.1001-7216.2020.9056
• Experimental Technique • Previous Articles
Mingxue CHEN1,2,#, Huan YANG2,#, Zhaoyun CAO2, Youning MA2, Zhenzhen CAO2, Fangmin CHENG1,*( )
)
Received:2019-05-15
Revised:2019-08-01
Online:2020-05-15
Published:2020-05-10
Contact:
Fangmin CHENG
About author:#These authors contributed equally to this work
陈铭学1,2,#, 杨欢2,#, 曹赵云2, 马有宁2, 曹珍珍2, 程方民1,*( )
)
通讯作者:
程方民
作者简介:#共同第一作者
基金资助:CLC Number:
Mingxue CHEN, Huan YANG, Zhaoyun CAO, Youning MA, Zhenzhen CAO, Fangmin CHENG. Absolute Quantification of Proteins in Rice Leaf Based on Multiple Reaction Monitoring Mass Spectrometry[J]. Chinese Journal OF Rice Science, 2020, 34(3): 278-286.
陈铭学, 杨欢, 曹赵云, 马有宁, 曹珍珍, 程方民. 基于多反应监测质谱技术的水稻叶片蛋白绝对定量方法[J]. 中国水稻科学, 2020, 34(3): 278-286.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2020.9056
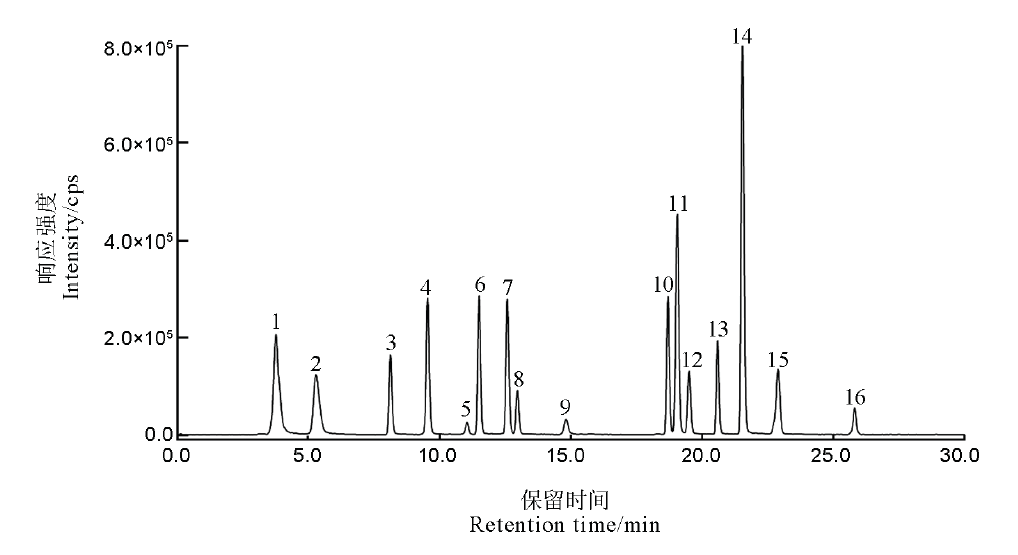
Fig. 1. Total ion chromatogram of 16 kinds of signature peptides. 1, P12085; 2, Q40677; 3, P0C2Z6; 4, Q8H8D6; 5, Q8GTK4; 6, A3BLC3; 7, Q650W6; 8, Q9ZP20; 9, Q0D5P8; 10, Q948T6; 11, Q8S718; 12, Q7X8A1; 13, Q0DEU8; 14, Q7XV11; 15, Q7XZW5; 16, Q69S39.
| 蛋白质序列号 Accession | 线性方程 Linear equation | 相关系数 r | 线性范围 Linear range/(nmol·L-1) | 检出限 LOD/(nmol·L-1) | 定量限 LOQ/(nmol·L-1) |
|---|---|---|---|---|---|
| Q40677 | y=1.97e4 x + 5.21e5 | 0.9965 | 0.5~250.0 | 0.17 | 0.5 |
| P12085 | y=1.75e4 x + 3.11e5 | 0.9990 | 0.6~300.0 | 0.20 | 0.6 |
| A3BLC3 | y=0.98e4 x + 5.47e5 | 0.9974 | 0.3~200.0 | 0.10 | 0.3 |
| Q9ZP20 | y=3.25e3 x + 2.94e5 | 0.9983 | 1.0~1000.0 | 0.33 | 1.0 |
| Q0D5P8 | y=4.14e3 x + 6.54e5 | 0.9909 | 2.5~1000.0 | 0.80 | 2.5 |
| Q7X8A1 | y=8.59e4 x + 2.95e6 | 0.9993 | 0.5~250.0 | 0.17 | 0.5 |
| Q0DEU8 | y=2.14e3 x + 4.87e5 | 0.9974 | 0.5~250.0 | 0.17 | 0.5 |
| Q7XV11 | y=4.59e3 x + 2.65e5 | 0.9986 | 0.1~100.0 | 0.03 | 0.1 |
| Q7XZW5 | y=2.56e3 x + 2.46e5 | 0.9959 | 0.5~250.0 | 0.17 | 0.5 |
| P0C2Z6 | y=9.13e3 x + 4.48e4 | 0.9967 | 0.7~300.0 | 0.23 | 0.7 |
| Q650W6 | y=7.54e3 x + 6.21e5 | 0.9987 | 0.4~200.0 | 0.13 | 0.4 |
| Q69S39 | y=1.58e3 x + 4.64e5 | 0.9988 | 1.7~1000.0 | 0.56 | 1.7 |
| Q8GTK4 | y=2.51e4 x + 3.36e6 | 0.9989 | 1.8~1000.0 | 0.60 | 1.8 |
| Q8H8D6 | y=5.57e4 x + 3.78e5 | 0.9991 | 0.4~200.0 | 0.13 | 0.4 |
| Q8S718 | y=3.45e4 x + 2.49e5 | 0.9988 | 0.3~200.0 | 0.10 | 0.3 |
| Q948T6 | y=4.44e3 x + 4.25e5 | 0.9982 | 0.4~200.0 | 0.13 | 0.4 |
Table 1 Linear equations, correlation coeffcients(r), limits of detection(LOD) and limits of quantification (LOQ) of the 16 kinds of signature peptides.
| 蛋白质序列号 Accession | 线性方程 Linear equation | 相关系数 r | 线性范围 Linear range/(nmol·L-1) | 检出限 LOD/(nmol·L-1) | 定量限 LOQ/(nmol·L-1) |
|---|---|---|---|---|---|
| Q40677 | y=1.97e4 x + 5.21e5 | 0.9965 | 0.5~250.0 | 0.17 | 0.5 |
| P12085 | y=1.75e4 x + 3.11e5 | 0.9990 | 0.6~300.0 | 0.20 | 0.6 |
| A3BLC3 | y=0.98e4 x + 5.47e5 | 0.9974 | 0.3~200.0 | 0.10 | 0.3 |
| Q9ZP20 | y=3.25e3 x + 2.94e5 | 0.9983 | 1.0~1000.0 | 0.33 | 1.0 |
| Q0D5P8 | y=4.14e3 x + 6.54e5 | 0.9909 | 2.5~1000.0 | 0.80 | 2.5 |
| Q7X8A1 | y=8.59e4 x + 2.95e6 | 0.9993 | 0.5~250.0 | 0.17 | 0.5 |
| Q0DEU8 | y=2.14e3 x + 4.87e5 | 0.9974 | 0.5~250.0 | 0.17 | 0.5 |
| Q7XV11 | y=4.59e3 x + 2.65e5 | 0.9986 | 0.1~100.0 | 0.03 | 0.1 |
| Q7XZW5 | y=2.56e3 x + 2.46e5 | 0.9959 | 0.5~250.0 | 0.17 | 0.5 |
| P0C2Z6 | y=9.13e3 x + 4.48e4 | 0.9967 | 0.7~300.0 | 0.23 | 0.7 |
| Q650W6 | y=7.54e3 x + 6.21e5 | 0.9987 | 0.4~200.0 | 0.13 | 0.4 |
| Q69S39 | y=1.58e3 x + 4.64e5 | 0.9988 | 1.7~1000.0 | 0.56 | 1.7 |
| Q8GTK4 | y=2.51e4 x + 3.36e6 | 0.9989 | 1.8~1000.0 | 0.60 | 1.8 |
| Q8H8D6 | y=5.57e4 x + 3.78e5 | 0.9991 | 0.4~200.0 | 0.13 | 0.4 |
| Q8S718 | y=3.45e4 x + 2.49e5 | 0.9988 | 0.3~200.0 | 0.10 | 0.3 |
| Q948T6 | y=4.44e3 x + 4.25e5 | 0.9982 | 0.4~200.0 | 0.13 | 0.4 |
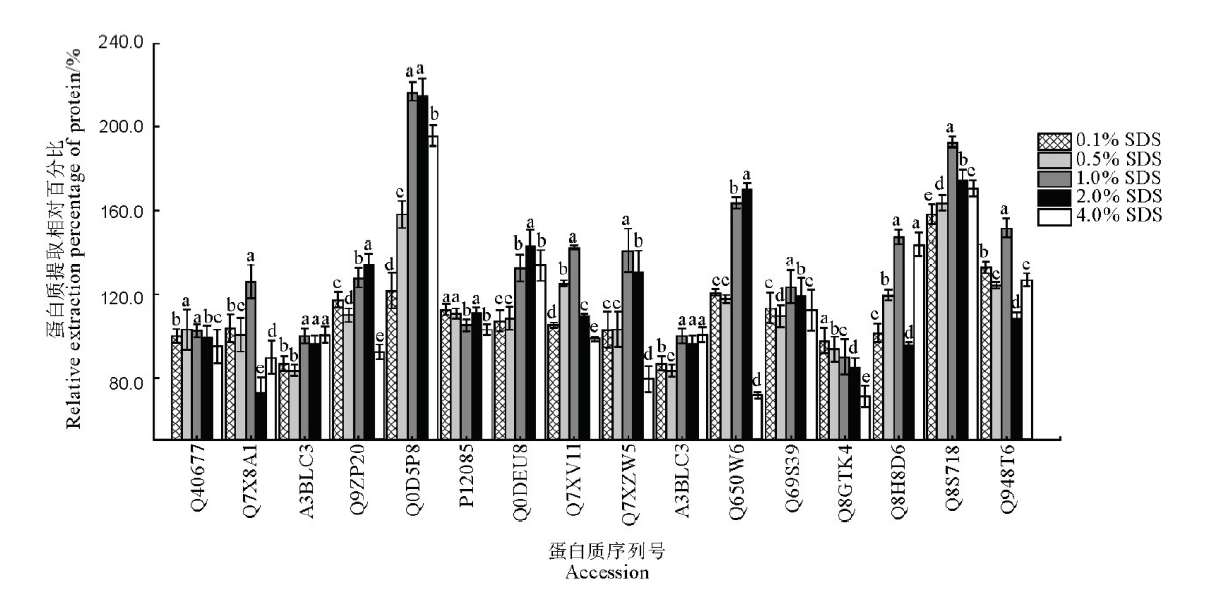
Fig. 2. Influence of different concentrations of SDS in extraction buffer on the extraction effects of 16 kinds of proteins in rice leaves. The data in the figure are mean ± SD (n=3), and different letters indicate significant differences at P<0.05, while the same letters mean no significant difference. The ordinate is the relative extraction percentage of protein with the SDS-free extraction amount of each target protein as 100%.
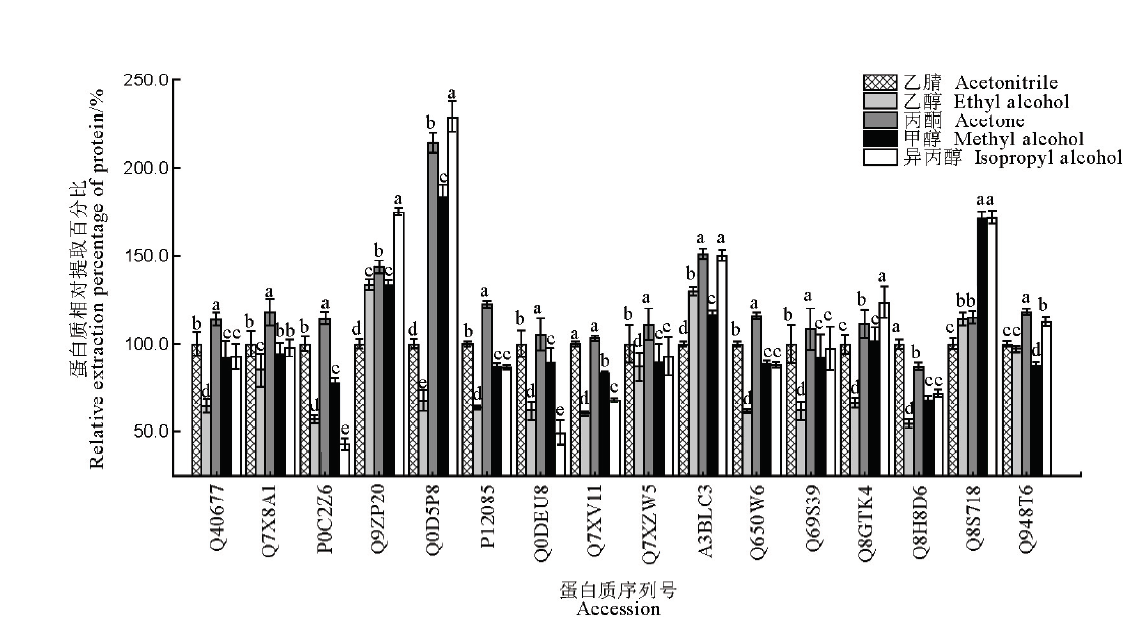
Fig. 3. Comparison of precipitation effects of 16 kinds of proteins in rice leaves among five different organic reagents. The data in the figure are mean ± SD (n=3), and different letters indicate significant differences at P<0.05, while the same letters mean no significant difference. The ordinate is the relative extraction percentage of target protein using various precipitation reagents, with acetonitrile-precipitated protein amount as 100%.
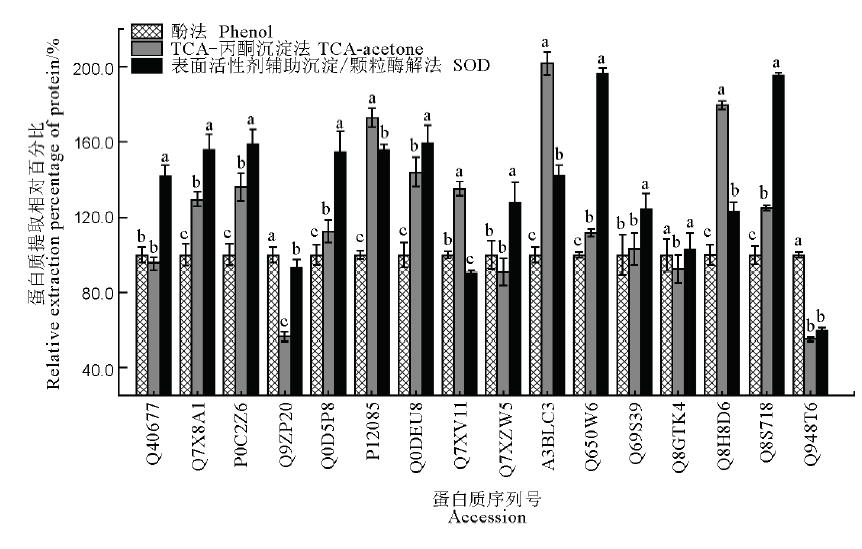
Fig. 4. Comparison of extraction effects of 16 proteins in rice leaves by SOD, phenol and TCA-acetone precipitation methods. The data in the figure are as mean ± SD (n=3), and different letters indicate significant differences at P<0.05, while the same letters mean no significant difference. The ordinate is the relative extraction percentage of target protein by various methods, with protein amount extracted by phenol method as 100%.
| 蛋白序列号Accession | 测定含量 Measured content/(μg·g-1) | 相对标准偏差 RSD/% |
|---|---|---|
| Q40677 | 589.5±57.5 | 9.8 |
| P12085 | 28.5±2.3 | 8.2 |
| A3BLC3 | 43.3±3.9 | 8.9 |
| Q9ZP20 | 50.2±3.6 | 7.1 |
| Q0D5P8 | 606.9±56.0 | 9.2 |
| Q7X8A1 | 328.3±27.1 | 8.3 |
| Q0DEU8 | 285.8±30.1 | 10.5 |
| Q7XV11 | 20.6±1.7 | 8.2 |
| Q7XZW5 | 153.2±14.0 | 9.1 |
| P0C2Z6 | 67.9±4.8 | 7.0 |
| Q650W6 | 25.5±2.9 | 11.3 |
| Q69S39 | 2818.1±219.9 | 7.8 |
| Q8GTK4 | 109.6±8.1 | 7.4 |
| Q8H8D6 | 6.0±0.4 | 6.2 |
| Q8S718 | 11.7±0.9 | 7.4 |
| Q948T6 | 11.8±1.7 | 13.9 |
Table 2 Determination of absolute contents of 16 kinds of proteins in rice leaves by SOD method(n=6).
| 蛋白序列号Accession | 测定含量 Measured content/(μg·g-1) | 相对标准偏差 RSD/% |
|---|---|---|
| Q40677 | 589.5±57.5 | 9.8 |
| P12085 | 28.5±2.3 | 8.2 |
| A3BLC3 | 43.3±3.9 | 8.9 |
| Q9ZP20 | 50.2±3.6 | 7.1 |
| Q0D5P8 | 606.9±56.0 | 9.2 |
| Q7X8A1 | 328.3±27.1 | 8.3 |
| Q0DEU8 | 285.8±30.1 | 10.5 |
| Q7XV11 | 20.6±1.7 | 8.2 |
| Q7XZW5 | 153.2±14.0 | 9.1 |
| P0C2Z6 | 67.9±4.8 | 7.0 |
| Q650W6 | 25.5±2.9 | 11.3 |
| Q69S39 | 2818.1±219.9 | 7.8 |
| Q8GTK4 | 109.6±8.1 | 7.4 |
| Q8H8D6 | 6.0±0.4 | 6.2 |
| Q8S718 | 11.7±0.9 | 7.4 |
| Q948T6 | 11.8±1.7 | 13.9 |
| [1] | 钱小红. 定量蛋白质组学分析方法[J]. 色谱, 2013, 31(8): 719-723. |
| Qian X H.Quantitative proteomics analysis[J]. Chinese Journal of Chromatography, 2013, 31(8): 719-723. (in Chinese with English abstract) | |
| [2] | 齐林玉, 姜淼, 郭艳, 王迎, 陈平. 质谱MRM技术结合稳定同位素标记法的应用进展[J]. 生命科学研究, 2015, 19(5): 444-451. |
| Qi L Y, Jiang M, Guo Y, Wang Y, Chen P.Progresses on application of MS MRM technology combining with stable isotope labeling[J]. Life Science Research, 2015, 19(5): 444-451. (in Chinese with English abstract) | |
| [3] | Veronika V, Zdenek S.A review on mass spectrometry-based quantitative proteomics: Targeted and data independent acquisition[J]. Analytica Chimica Acta, 2017, 964: 7-23. |
| [4] | 王素兰, 高华萍, 张菁, 叶翔. 基于稳定同位素标记和平行反应监测的蛋白质组学定量技术用于肝癌生物标志物的筛选和验证[J]. 色谱, 2017, 35(9): 934-940. |
| Wang S L, Gao H P, Zhang J, Ye X.Stable isotope labeling and parallel reaction monitoring-based proteomic quantification for biomarker screening and validation of hepatocellular carcinoma[J]. Chinese Journal of Chromatography, 2017, 35(9): 934-940. (in Chinese with English abstract) | |
| [5] | Chen Y T, Chen H W, Wu C F, Chu L J, Chiang W F, Wu C C, Yu J S, Tsai C H, Liang K H, Chang Y S, Wu M, Ou Yang W T. Development of a multiplexed liquid chromatography multiple-reaction-monitoring mass spectrometry (LC-MRM/MS) method for evaluation of salivary proteins as oral cancer biomarkers[J]. Molecular Cellular Proteomics, 2017, 16(5): 799-811. |
| [6] | Zhang J, Lai S, Cai Z, Chen Q, Huang B, Ren Y.Determination of bovine lactoferrin in dairy products by ultra-high performance liquid chromatography-tandem mass spectrometry based on tryptic signature peptides employing an isotope-labeled winged peptide as internal standard[J]. Analytica Chimica Acta, 2014, 4(829): 33-39. |
| [7] | Planque M, Arnould T, Delahaut P, Renardb P, Dieub M, Gillard N.Development of a strategy for the quantification of food allergens in several food products by mass spectrometry in a routine laboratory[J]. Food Chemistry, 2019, 274: 35-45. |
| [8] | Rautengarten C, Ebert B, Heazlewood J L.Absolute quantitation of in vitro expressed plant membrane proteins by targeted proteomics (MRM) for the determination of kinetic parameters[J]. Methods in Molecular Biology, 2018, 1696: 217-234. |
| [9] | Gong C, Zheng N, Zeng J, Aubry A F, Arnold M E.Post-pellet-digestion precipitation and solid phase extraction: A practical and efficient workflow to extract surrogate peptides for ultra-high performance liquid chromatography-tandem mass spectrometry bioanalysis of a therapeutic antibody in the low ng/mL range[J]. Journal of Chromatography A, 2015, 1424: 27-36. |
| [10] | Wu X, Xiong E, Wang W, Scali M, Cresti M.Universal sample preparation method integrating trichloroacetic acid/acetone precipitation with phenol extraction for crop proteomic analysis[J]. Nature Protocols, 2014, 9(2): 362-374. |
| [11] | Luís I M, Alexandre B M, Oliveira M M, Abreu I A.Selection of an appropriate protein extraction method to study the phosphoproteome of maize photosynthetic tissue[J/OL].PLoS One, 2016, 11(10): e0164387. |
| [12] | Kilambi H V, Manda K, Sanivarapu H, Maurya V K, Sharma R, Sreelakshmi Y.Shotgun proteomics of tomato fruits: Evaluation, optimization and validation of sample preparation methods and mass spectrometric parameters [J/OL]. Frontiers in Plant Science, 2016, 7: 969. |
| [13] | Bereman M S, Egertson J D, MacCoss M J. Comparison between procedures using SDS for shotgun proteomic analyses of complex samples[J]. Proteomics, 2011, 11(14): 2931-5. |
| [14] | Yang L, Ji J, Harris-Shultz K R, Wang H, Wang H, Abd-Allah E F, Luo Y, Hu X Y. The dynamic changes of the plasma membrane proteins and the protective roles of nitric oxide in rice subjected to heavy metal cadmium stress[J/OL].Frontiers in Plant Science, 2016, 7: 190. |
| [15] | 高欢欢, 马有宁, 林晓燕, 柴爽爽, 秦美玲, 张涵彤, 何巧, 陈铭学. 亲水作用-反相二维液相色谱串联质谱法鉴定水稻蛋白质[J]. 分析化学, 2018, 46(5): 650-657. |
| Gao H H, Ma Y N, Lin X Y, Chai S S, Qin M L, Zhang H T, He Q, ChenM X. Development of two-dimensional liquid chromatography coupled with tandem mass spectrometry for identification of extracted proteins of rice leaves: Hydrophilic interaction-reversed-phase approach[J]. Chinese Journal of Analytical Chemistry, 2018, 46(5): 650-657. (in Chinese with English abstract) | |
| [16] | Ippoushi K, Sasanuma M, Oike H, Kobori M, Maeda-Yamamoto M.Absolute quantification of protein NP24 in tomato fruit by liquid chromatography/tandem mass spectrometry using stable isotope-labelled tryptic peptide standard[J]. Food Chemistry, 2015, 173: 238-242. |
| [17] | Wisniewski J R, Zouqman A, Nagaraj N, Mann M.Universal sample preparation method for proteome analysis[J]. Nature Methods, 2009, 6(5): 359-362. |
| [18] | Bru-Martínez R, Martínez-Márquez A, Morante-Carriel J, Sellés-Marchart S, Martínez-Esteso M J, Pineda-Lucas J L, Luque I. Targeted quantification of isoforms of a thylakoid-bound protein: MRM method development[J]. Methods in Molecular Biology, 2018, 1696: 147-162. |
| [19] | An B, Zhang M, Johnson R W, Qu J.Surfactant-aided precipitation/on-pellet-digestion (SOD) procedure provides robust and rapid sample preparation for reproducible, accurate and sensitive LC/MS quantification of therapeutic protein in plasma and tissues[J]. Analytical Chemistry, 2015, 87(7): 4023-4029. |
| [20] | Santa C, Anjo S I, Manadas B.Protein precipitation of diluted samples in SDS-containing buffer with acetone leads to higher protein recovery and reproducibility in comparison with TCA/acetone approach[J]. Proteomics, 2016, 16(13): 1847-1851. |
| [21] | Botelho D, Wall M J, Vieira D B, Fitzsimmons S, Liu F, Doucette A.Top-down and bottom-up proteomics of SDS containing solutions following mass-based separation[J]. Journal of Proteome Research, 2010, 9(6): 2863-2870. |
| [22] | Zhang N, Chen R, Young N, Wishart D, Winter P, Weiner J H, Li L.Comparison of SDS and methanol-assisted protein solubilization and digestion methods for Escherichia coli membrane proteome analysis by 2D LC-MS/MS[J]. Proteomics, 2007, 7(4): 484-493. |
| [23] | Ilavenil S, Al-Dhabi N A, Srigopalram S, Kim Y O, Agastian P, BaaruKi R, Choon Choi K C, Arasu M V, Park C G, Park K H. Removal of SDS from biological protein digests for proteomic analysis by mass spectrometry[J]. Proteome Science, 2016, 14(1): 11. |
| [24] | Ouyang Z, Furlong M T, Wu S, Sleczka B, Tamura J, Wang H, Suchard S, Suri A, Olah T, Tymiak A, Jemal M.Pellet digestion: A simple and efficient sample preparation technique for LC-MS/MS quantification of large therapeutic proteinsin plasma[J]. Bioanalysis, 2012, 4(1): 17-28. |
| [25] | Srivastava A K. A Comparison of protein extraction methods using organic solvents for secretome of aspergillus fumigatus strain (MTCC 1811). International Journal of Scientific Research, 2015, 4(6): 2277-8179. |
| [26] | 张冬梅, 马慧萍, 贾正平. 纳升级反相液相色谱串联质谱法分析海马蛋白质组[J]. 药物分析杂志, 2018, 38(1): 118-123. |
| Zhang D M, Ma H P, Jia Z P.Proteomic analysis of Hippocampus using nanoflow reversed phase liquid chromatography-tandem mass spectrometry[J]. Chinese Journal of Pharmaceutical Analysis, 2018, 38(1): 118-123. (in Chinese with English abstract) | |
| [27] | Rezeli M, Sjödin K, Lindberg H, Gidlöf O, Lindahl B, Jernberg T, Spaak J, Erlinge D, Marko-Varga G.Quantitation of 87 proteins by nLC-MRM/MS in human plasma: workflow for large-scale analysis of biobank samples[J]. Journal of Proteome Research, 2017, 16(19): 3242-3254. |
| [28] | Hoofnagle A N, Becker J O, Oda M N, Cavigiolio G, Mayer P, Vaisar T.Multiple-reaction monitoring-mass spectrometric assays can accurately measure the relative protein abundance in complex mixture[J]. Clinical Chemistry, 2012, 58(4): 777-781. |
| [1] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [2] | TONG Qi, WANG Chunyan, QUE Yawei, XIAO Yu, WANG Zhengyi. Identification and Functional Characterization of the Heat Shock Protein (HSP) 40 Encoding Gene, MoMHF6, in Magnaporthe oryzae [J]. Chinese Journal OF Rice Science, 2023, 37(6): 563-576. |
| [3] | QI Panpan, GUO Liuming, LI Jing, LÜ Mingfang, YUAN Zhengjie, ZHANG Hengmu. cDNA Cloning and Molecular Characterization of OsTAF12b Gene in Oryza sativa [J]. Chinese Journal OF Rice Science, 2023, 37(6): 577-586. |
| [4] | HAN Cong, HE Yuchang, WU Lijuan, JIA Lili, WANG Lei, E Zhiguo. Research Progress in the Function of Basic Leucine Zipper (bZIP) Protein Family in Rice [J]. Chinese Journal OF Rice Science, 2023, 37(4): 436-448. |
| [5] | LU Dandan, YONG Mingling, TAO Yu, YE Miao, ZHANG Zujian. Characteristics of Grain Protein Accumulation and Its Response to Nitrogen Level in Good Taste Rice Varieties [J]. Chinese Journal OF Rice Science, 2022, 36(5): 520-530. |
| [6] | WANG Mengjie, WU Junnan, LIU Yaobin, XU Chunchun, FENG Jinfei, LI Fengbo, FANG Fuping. Effect of Feed Protein Contents on Gaseous Nitrogen Emissions, Growth Performance of Catfish and Nitrogen Utilization in Rice-yellow Catfish Co-culture Model [J]. Chinese Journal OF Rice Science, 2022, 36(4): 428-438. |
| [7] | Kai LU, Tao CHEN, Shu YAO, Wenhua LIANG, Xiaodong WEI, Yadong ZHANG, Cailin WANG. Functional Analysis on Four Receptor-like Protein Kinases Under Salt Stress in Rice [J]. Chinese Journal OF Rice Science, 2021, 35(2): 103-111. |
| [8] | Zhidong WANG, Yibo CHEN, Rong GONG, Shaochuan ZHOU, Chongrong WANG, Hong LI, Daoqiang HUANG, Degui ZHOU, Lei ZHAO, Yangyang PAN, Yiqiang YANG, LIXiaofang. Correlation Between SPAD Value of Flag Leaf and Rice Quality of High Quality indica Rice [J]. Chinese Journal OF Rice Science, 2021, 35(1): 89-97. |
| [9] | Xiaohan TANG, Shijia LIU, Xi LIU, Yunlu TIAN, Yunlong WANG, Xuan TENG, Erchao DUAN, Yuanyan ZHANG, Ling JIANG, Wenwei ZHANG, Yihua WANG, Jianmin WAN. Tryptophanyl-tRNA Synthetase Gene WRS1 Regulates Rice Seed Development [J]. Chinese Journal OF Rice Science, 2020, 34(5): 383-396. |
| [10] | Lei PAN, Lihua WANG, Feng ZHU, Yangchun HAN, Pei WANG, Jichao FANG. Expression Profiles and Functions of Small Heat Shock Proteins in Nilaparvata lugens [J]. Chinese Journal OF Rice Science, 2020, 34(1): 37-45. |
| [11] | Lü SHI, Xinyue ZHANG, Huiyan SUN, Xianmei CAO, Jian LIU, Zujian ZHANG. Relationship of Grain Protein Content with Cooking and Eating Quality as Affected by Nitrogen Fertilizer at Late Growth Stage for Different Types of Rice Varieties [J]. Chinese Journal OF Rice Science, 2019, 33(6): 541-552. |
| [12] | Hui FENG, Yalei FAN, Jinfeng ZHANG, Hongli ZHU, Lihui WEI. Protective Effect of Glutaredoxin (AbGrx-1) on Aphelenchoides besseyi Under Oxidative Stress [J]. Chinese Journal OF Rice Science, 2019, 33(6): 565-574. |
| [13] | Jing HE, Han LIU, Liuming GUO, Jing LI, Hengmu ZHANG. Characterization and Application of Antibodies Against a Small Heat Shock Protein OsHSP20 from Oryza sativa [J]. Chinese Journal OF Rice Science, 2019, 33(3): 235-240. |
| [14] | Jie ZHANG, Baijie WAN, Pengxiang SHANG, Xinlun DING, Zhenguo DU, Zujian WU. Expression of Pns6 and P8 Proteins of Rice Dwarf Virus After Virus Infecting Rice Protoplasts [J]. Chinese Journal OF Rice Science, 2019, 33(2): 118-123. |
| [15] | Qiaoli CHEN, Feng WANG, Danlei LI, Yaming LING, Ruizhi ZHANG. Expression Under Hypertonic Osmotic Stress of Ab-lea from Aphelenchoides besseyi [J]. Chinese Journal OF Rice Science, 2017, 31(6): 652-657. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||