
Chinese Journal OF Rice Science ›› 2020, Vol. 34 ›› Issue (3): 245-255.DOI: 10.16819/j.1001-7216.2020.9126
• Research Papers • Previous Articles Next Articles
Zhaomeng XU1,2, Lihua LI2,3, Xiaoqing GAO1, Zhengjie YUAN2, Xin LI2, Xudan TIAN1,2, Lanlan WANG2, Shaohong QU2,*( )
)
Received:2019-11-23
Revised:2020-03-11
Online:2020-05-15
Published:2020-05-10
Contact:
Shaohong QU
许赵蒙1,2, 李利华2,3, 高晓庆1, 袁正杰2, 李莘2, 田旭丹1,2, 王岚岚2, 瞿绍洪2,*( )
)
通讯作者:
瞿绍洪
基金资助:CLC Number:
Zhaomeng XU, Lihua LI, Xiaoqing GAO, Zhengjie YUAN, Xin LI, Xudan TIAN, Lanlan WANG, Shaohong QU. Comparative Transcriptome Analysis of Transgenic Rice Line Carrying the Rice Blast Resistance Gene Pi9[J]. Chinese Journal OF Rice Science, 2020, 34(3): 245-255.
许赵蒙, 李利华, 高晓庆, 袁正杰, 李莘, 田旭丹, 王岚岚, 瞿绍洪. 转Pi9抗稻瘟病基因水稻株系的比较转录组分析[J]. 中国水稻科学, 2020, 34(3): 245-255.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2020.9126
| 基因 Gene | 正向引物 Forward primer(5' → 3') | 反向引物 Reverse primer(5' → 3') |
|---|---|---|
| Os03g08020 | CCACGGGCCATCTGATCTAC | AGTCAATGATGAGCACGGCA |
| Os04g41620 | CTTCTTCGCGCATGTCACAC | GGTGCACGTTGTTCATCCAG |
| Os10g39680 | ATTGACAGGGCAATCGAACT | GCCGCCGTTGATGATGTTG |
| Os08g41100 | AACTGGTGGTCCATTGGCTT | TCATGGGCTGATGGTTGCTT |
| Os03g06520 | ATCACCCGCTTCTTCAGCAA | CAAAGCTCCTTCCTACGGCA |
| Os03g09930 | GTGAACTTCGCTGCTGGTTG | TGAGGGATCAGTACTGCAACTG |
| Os04g55800 | CAAATGTGGCCTGAATCGCC | CCAGCATTGTGGTTCACAGC |
| Os06g05160 | ATGCACTCCGTCTTCAAGCA | GAGCCACTGAAGGTCAGCAT |
| Os09g06499 | CAAAGTTGGCTGGGCTTCAC | TAAGCCAACCGAGCCTTAGC |
| Os01g58280 | TGTCCCTCTCACTTGGTGTTG | CCCTTGAACAATTGCCACCA |
| Os03g03550 | CAACGAGCTCAAGTTCAGGC | TGCACCGTTCTGTTGTGACT |
| Os06g14190 | TGGTGGTAGAGCTTCATGTGG | AGTGGTACCCTCCTCCCATC |
| Os04g27670 | GCAGTCATAGCATGCGCAAA | GTGTGGCAAATGTGTAGGCA |
| Os05g31040 | TCCTATCCTCAGCACTTGGC | GGTAGTGACGCTGATGCCTT |
| Os05g41290 | AGGACCCTGAAAAATGGGACA | TTGCTTTTGCAGCAAGTGGG |
| Os08g07340 | GTGGGGCTTTTGTTTCGGTC | GACATGCATGGCAACTTCGG |
| Os10g03850 | TCCCGAGGGTTTCACACCTA | GCAAGAAGAAACAACACCATGC |
Table 1 Primers used for real-time quantitative reverse-transcription PCR.
| 基因 Gene | 正向引物 Forward primer(5' → 3') | 反向引物 Reverse primer(5' → 3') |
|---|---|---|
| Os03g08020 | CCACGGGCCATCTGATCTAC | AGTCAATGATGAGCACGGCA |
| Os04g41620 | CTTCTTCGCGCATGTCACAC | GGTGCACGTTGTTCATCCAG |
| Os10g39680 | ATTGACAGGGCAATCGAACT | GCCGCCGTTGATGATGTTG |
| Os08g41100 | AACTGGTGGTCCATTGGCTT | TCATGGGCTGATGGTTGCTT |
| Os03g06520 | ATCACCCGCTTCTTCAGCAA | CAAAGCTCCTTCCTACGGCA |
| Os03g09930 | GTGAACTTCGCTGCTGGTTG | TGAGGGATCAGTACTGCAACTG |
| Os04g55800 | CAAATGTGGCCTGAATCGCC | CCAGCATTGTGGTTCACAGC |
| Os06g05160 | ATGCACTCCGTCTTCAAGCA | GAGCCACTGAAGGTCAGCAT |
| Os09g06499 | CAAAGTTGGCTGGGCTTCAC | TAAGCCAACCGAGCCTTAGC |
| Os01g58280 | TGTCCCTCTCACTTGGTGTTG | CCCTTGAACAATTGCCACCA |
| Os03g03550 | CAACGAGCTCAAGTTCAGGC | TGCACCGTTCTGTTGTGACT |
| Os06g14190 | TGGTGGTAGAGCTTCATGTGG | AGTGGTACCCTCCTCCCATC |
| Os04g27670 | GCAGTCATAGCATGCGCAAA | GTGTGGCAAATGTGTAGGCA |
| Os05g31040 | TCCTATCCTCAGCACTTGGC | GGTAGTGACGCTGATGCCTT |
| Os05g41290 | AGGACCCTGAAAAATGGGACA | TTGCTTTTGCAGCAAGTGGG |
| Os08g07340 | GTGGGGCTTTTGTTTCGGTC | GACATGCATGGCAACTTCGG |
| Os10g03850 | TCCCGAGGGTTTCACACCTA | GCAAGAAGAAACAACACCATGC |
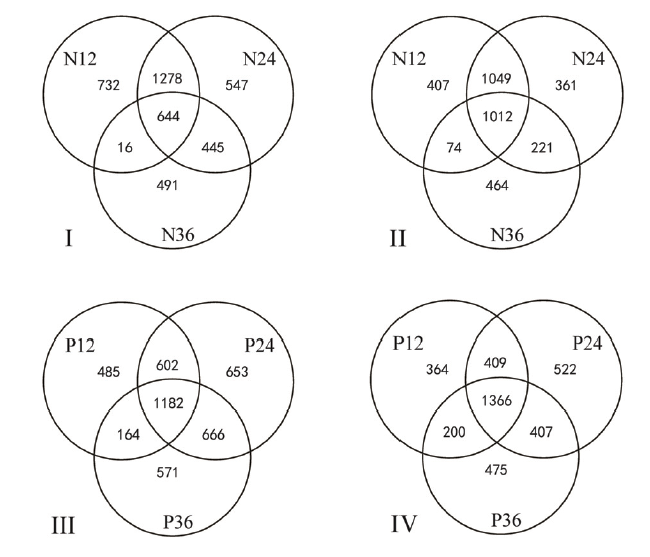
Fig. 1. Number of differentially expressed genes(DEGs) in NPB/Pi9 and NPB after inoculation. I and III, Up-regulated genes; II and IV, Down-regulated genes; N12, N24 and N36, The numbers of DEGs at 12 h, 24 h and 36 h after inoculation in NPB; P12, P24 and P36, The numbers of DEGs at 12 h, 24 h and 36 h after inoculation in NPB/Pi9.
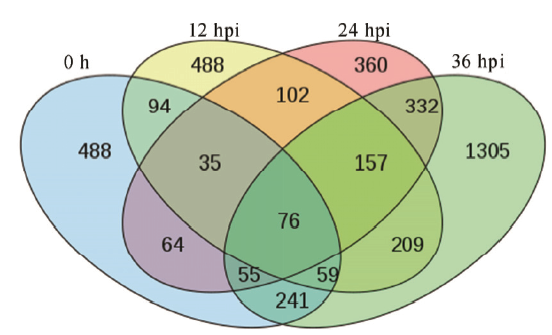
Fig. 4. Venn diagram of differentially expressed genes (DEGs) between the resistant genotype NPB/Pi9 and the susceptible genotype NPB. Each color represents a set of differential comparison, and the numbers represent the gene numbers that are unique or shared by different experiments. An overlapping region is the genes shared by different groups, and a non-overlapping region is the gene unique to each group. 0 h, 12 hpi, 24 hpi and 36 hpi represent 0, 12, 24 and 36 hours after inoculation, respectively.
| 接种后时间 Hours after inoculation /h | 生物学过程 Biological process | 分子功能 Molecular function | 细胞组分 Cellular component |
|---|---|---|---|
| 0 | RNA代谢RNA metabolic process (GO: 0016070) | 核酸酶活性Nuclease activity (GO: 0004518) | 类囊体膜Thylakoid membrane (GO: 0042651) |
| 核酸代谢Nucleic acid metabolic process (GO: 0090304) | 锌离子结合Zinc ion binding (GO: 0008270) | 细胞外区域Extracellular region (GO: 0005576) | |
| 转录调控Regulation of transcription (GO: 0006355) | 转录调控因子活性Transcription regulator activity(GO: 0140110) | 包膜Envelope (GO: 0031975) | |
| 氧化还原过程Oxidation-reduction process(GO: 0055114) | 氧化还原酶活性Oxidoreductase activity(GO: 0016491) | 叶绿体组分Chloroplast part (GO: 0044434) | |
| 对刺激的应答Response to stimulus (GO: 0050896) | 转移酶活性Transferase activity (GO: 0016740) | 细胞器组分Organelle part (GO: 0044422) | |
| 12 | 对刺激的应答Response to stimulus (GO: 0050896) | 铁离子结合Iron ion binding (GO: 0005506) | 叶绿体组分Chloroplast part (GO: 0044434) |
| 光合作用-捕光Photosynthesis-light harvesting (GO: 0009765) | 氧化还原酶活性Oxidoreductase activity (GO: 0016491) | 捕光复合体Light-harvesting complex(GO: 0030076) | |
| 次生代谢过程Secondary metabolic process (GO: 0019748) | 转录调控因子活性Transcription regulator activity(GO: 0140110) | 类囊体膜Thylakoid membrane (GO: 0042651) | |
| 转录调控Regulation of transcription (GO: 0006355) | 转移酶活性Transferase activity (GO: 0016740) | 包膜Envelope (GO: 0031975) | |
| 不饱和脂肪酸代谢过程Unsaturated fatty acid metabolic process(GO: 0033559) | 催化活性Catalytic activity (GO: 0003824) | 细胞组分Cell part (GO: 0044464) | |
| 24 | 光合作用Photosynthesis (GO: 0015979) | 铁离子结合Iron ion binding (GO: 0005506) | 光系统Photosystem (GO: 0009521) |
| 次级代谢生物合成Secondary metabolite biosynthetic process (GO: 0044550) | 转录调控因子活性Transcription regulator activity(GO: 0140110) | 类囊体膜Thylakoid membrane (GO: 0042651) | |
| 对刺激的应答Response to stimulus (GO: 0050896) | 激酶活性Kinase activity (GO: 0016301) | 细胞外区域Extracellular region (GO: 0005576) | |
| 对激素的反应Response to hormone (GO: 0009725) | 氧化还原酶活性Oxidoreductase activity (GO: 0016491) | 膜组分Membrane part (GO: 0044425) | |
| 碳水化合物代谢Carbohydrate metabolic process (GO: 0005975) | 催化活性Catalytic activity (GO: 0003824) | 叶绿体Chloroplast (GO: 0009507) | |
| 36 | 光合作用Photosynthesis (GO: 0015979) | 氧化还原酶活性Oxidoreductase activity (GO: 0016491) | 光系统Photosystem (GO: 0009521) |
| 对激素的反应Response to hormone (GO: 0009725) | 过氧化物酶活性Peroxidase activity (GO: 0004601) | 类囊体膜Thylakoid membrane (GO: 0042651) | |
| 氧化还原过程Oxidation-reduction process (GO: 0055114) | 离子结合Ion binding (GO: 0043167) | 细胞外区域Extracellular region (GO: 0005576) | |
| 信号转导Signal transduction (GO: 0007165) | 转移酶活性Transferase activity (GO: 0016740) | 膜结合细胞器Membrane-bounded organelle(GO: 0043227) | |
| 次级代谢生物合成Secondary metabolite biosynthetic process (GO: 0044550) | 水解酶活性Hydrolase activity (GO: 0016787) | 叶绿体Chloroplast (GO: 0009507) |
Table 2 GO enrichment of the differentially expressed genes between NPB/Pi9 and NPB.
| 接种后时间 Hours after inoculation /h | 生物学过程 Biological process | 分子功能 Molecular function | 细胞组分 Cellular component |
|---|---|---|---|
| 0 | RNA代谢RNA metabolic process (GO: 0016070) | 核酸酶活性Nuclease activity (GO: 0004518) | 类囊体膜Thylakoid membrane (GO: 0042651) |
| 核酸代谢Nucleic acid metabolic process (GO: 0090304) | 锌离子结合Zinc ion binding (GO: 0008270) | 细胞外区域Extracellular region (GO: 0005576) | |
| 转录调控Regulation of transcription (GO: 0006355) | 转录调控因子活性Transcription regulator activity(GO: 0140110) | 包膜Envelope (GO: 0031975) | |
| 氧化还原过程Oxidation-reduction process(GO: 0055114) | 氧化还原酶活性Oxidoreductase activity(GO: 0016491) | 叶绿体组分Chloroplast part (GO: 0044434) | |
| 对刺激的应答Response to stimulus (GO: 0050896) | 转移酶活性Transferase activity (GO: 0016740) | 细胞器组分Organelle part (GO: 0044422) | |
| 12 | 对刺激的应答Response to stimulus (GO: 0050896) | 铁离子结合Iron ion binding (GO: 0005506) | 叶绿体组分Chloroplast part (GO: 0044434) |
| 光合作用-捕光Photosynthesis-light harvesting (GO: 0009765) | 氧化还原酶活性Oxidoreductase activity (GO: 0016491) | 捕光复合体Light-harvesting complex(GO: 0030076) | |
| 次生代谢过程Secondary metabolic process (GO: 0019748) | 转录调控因子活性Transcription regulator activity(GO: 0140110) | 类囊体膜Thylakoid membrane (GO: 0042651) | |
| 转录调控Regulation of transcription (GO: 0006355) | 转移酶活性Transferase activity (GO: 0016740) | 包膜Envelope (GO: 0031975) | |
| 不饱和脂肪酸代谢过程Unsaturated fatty acid metabolic process(GO: 0033559) | 催化活性Catalytic activity (GO: 0003824) | 细胞组分Cell part (GO: 0044464) | |
| 24 | 光合作用Photosynthesis (GO: 0015979) | 铁离子结合Iron ion binding (GO: 0005506) | 光系统Photosystem (GO: 0009521) |
| 次级代谢生物合成Secondary metabolite biosynthetic process (GO: 0044550) | 转录调控因子活性Transcription regulator activity(GO: 0140110) | 类囊体膜Thylakoid membrane (GO: 0042651) | |
| 对刺激的应答Response to stimulus (GO: 0050896) | 激酶活性Kinase activity (GO: 0016301) | 细胞外区域Extracellular region (GO: 0005576) | |
| 对激素的反应Response to hormone (GO: 0009725) | 氧化还原酶活性Oxidoreductase activity (GO: 0016491) | 膜组分Membrane part (GO: 0044425) | |
| 碳水化合物代谢Carbohydrate metabolic process (GO: 0005975) | 催化活性Catalytic activity (GO: 0003824) | 叶绿体Chloroplast (GO: 0009507) | |
| 36 | 光合作用Photosynthesis (GO: 0015979) | 氧化还原酶活性Oxidoreductase activity (GO: 0016491) | 光系统Photosystem (GO: 0009521) |
| 对激素的反应Response to hormone (GO: 0009725) | 过氧化物酶活性Peroxidase activity (GO: 0004601) | 类囊体膜Thylakoid membrane (GO: 0042651) | |
| 氧化还原过程Oxidation-reduction process (GO: 0055114) | 离子结合Ion binding (GO: 0043167) | 细胞外区域Extracellular region (GO: 0005576) | |
| 信号转导Signal transduction (GO: 0007165) | 转移酶活性Transferase activity (GO: 0016740) | 膜结合细胞器Membrane-bounded organelle(GO: 0043227) | |
| 次级代谢生物合成Secondary metabolite biosynthetic process (GO: 0044550) | 水解酶活性Hydrolase activity (GO: 0016787) | 叶绿体Chloroplast (GO: 0009507) |
| 接种后时间 Hours after inoculation / h | KO 注释 KO ID | KEGG通路 KEGG pathway | 差异表达基因数目 No. of differentially expressed genes |
|---|---|---|---|
| 12 | KO00196 | 光合作用天线蛋白Photosynthesis-antenna proteins | 13 |
| KO00195 | 光合作用Photosynthesis | 15 | |
| 24 | KO00196 | 光合作用天线蛋白Photosynthesis-antenna proteins | 9 |
| KO00940 | 苯丙烷类生物合成Phenylpropanoid biosynthesis | 18 | |
| KO00710 | 光合生物碳素固定Carbon fixation in photosynthetic organisms | 11 | |
| KO00051 | 果糖和甘露糖代谢Fructose and mannose metabolism | 9 | |
| 36 | KO00940 | 苯丙烷类生物合成Phenylpropanoid biosynthesis | 42 |
| KO00196 | 光合作用天线蛋白Photosynthesis-antenna proteins | 13 | |
| KO00360 | 苯丙氨酸代谢Phenylalanine metabolism | 30 | |
| KO04075 | 植物激素信号转导Plant hormone signal transduction | 40 | |
| KO00941 | 类黄酮生物合成Flavonoid biosynthesis | 9 |
Table 3 KEGG enrichment of differentially expressed genes between NPB/Pi9 and NPB after M. oryzae inoculation.
| 接种后时间 Hours after inoculation / h | KO 注释 KO ID | KEGG通路 KEGG pathway | 差异表达基因数目 No. of differentially expressed genes |
|---|---|---|---|
| 12 | KO00196 | 光合作用天线蛋白Photosynthesis-antenna proteins | 13 |
| KO00195 | 光合作用Photosynthesis | 15 | |
| 24 | KO00196 | 光合作用天线蛋白Photosynthesis-antenna proteins | 9 |
| KO00940 | 苯丙烷类生物合成Phenylpropanoid biosynthesis | 18 | |
| KO00710 | 光合生物碳素固定Carbon fixation in photosynthetic organisms | 11 | |
| KO00051 | 果糖和甘露糖代谢Fructose and mannose metabolism | 9 | |
| 36 | KO00940 | 苯丙烷类生物合成Phenylpropanoid biosynthesis | 42 |
| KO00196 | 光合作用天线蛋白Photosynthesis-antenna proteins | 13 | |
| KO00360 | 苯丙氨酸代谢Phenylalanine metabolism | 30 | |
| KO04075 | 植物激素信号转导Plant hormone signal transduction | 40 | |
| KO00941 | 类黄酮生物合成Flavonoid biosynthesis | 9 |
| 差异表达基因类别 DEG category | 基因功能分类 Gene function | 差异表达基因数目DEG number | ||
|---|---|---|---|---|
| 接种后12 h 12 hpi | 接种后24 h 24 hpi | 接种后36 h 36 hpi | ||
| 抗病相关基因 Resistance-related genes | NBS-LRR | 12 | 10 | 23 |
| RPM1/RPP13 | 10 | 9 | 11 | |
| 其他Others | 3 | 2 | 4 | |
| 转录因子 Transcription factor | WRKY | 16 | 17 | 25 |
| MYB | 18 | 19 | 41 | |
| bZIP | 6 | 8 | 9 | |
| ERF/AP2 | 2 | 2 | 9 | |
| bHLH | 0 | 2 | 5 | |
| 其他Others | 14 | 9 | 25 | |
| 激酶Kinase | WAK | 8 | 10 | 12 |
| DUF26 | 2 | 1 | 13 | |
| MAPK | 1 | 3 | 9 | |
| 丝/苏氨酸蛋白激酶Serine/threonine-protein kinase | 4 | 10 | 19 | |
| 受体/类受体激酶Receptor/receptor-like kinase | 9 | 10 | 28 | |
| 钙/钙依赖性蛋白激酶 Calcium/calmodulin dependent protein kinases | 3 | 3 | 14 | |
| 凝集素样受体/蛋白激酶Lectin-like receptor kinase | 3 | 5 | 9 | |
| 蛋白激酶Protein kinase | 6 | 8 | 30 | |
| 其他激酶Other kinase | 15 | 25 | 34 | |
Table 4 DEGs related to kinases and transcription factors between NPB/Pi9 and NPB at various hours after inoculation(hpi).
| 差异表达基因类别 DEG category | 基因功能分类 Gene function | 差异表达基因数目DEG number | ||
|---|---|---|---|---|
| 接种后12 h 12 hpi | 接种后24 h 24 hpi | 接种后36 h 36 hpi | ||
| 抗病相关基因 Resistance-related genes | NBS-LRR | 12 | 10 | 23 |
| RPM1/RPP13 | 10 | 9 | 11 | |
| 其他Others | 3 | 2 | 4 | |
| 转录因子 Transcription factor | WRKY | 16 | 17 | 25 |
| MYB | 18 | 19 | 41 | |
| bZIP | 6 | 8 | 9 | |
| ERF/AP2 | 2 | 2 | 9 | |
| bHLH | 0 | 2 | 5 | |
| 其他Others | 14 | 9 | 25 | |
| 激酶Kinase | WAK | 8 | 10 | 12 |
| DUF26 | 2 | 1 | 13 | |
| MAPK | 1 | 3 | 9 | |
| 丝/苏氨酸蛋白激酶Serine/threonine-protein kinase | 4 | 10 | 19 | |
| 受体/类受体激酶Receptor/receptor-like kinase | 9 | 10 | 28 | |
| 钙/钙依赖性蛋白激酶 Calcium/calmodulin dependent protein kinases | 3 | 3 | 14 | |
| 凝集素样受体/蛋白激酶Lectin-like receptor kinase | 3 | 5 | 9 | |
| 蛋白激酶Protein kinase | 6 | 8 | 30 | |
| 其他激酶Other kinase | 15 | 25 | 34 | |
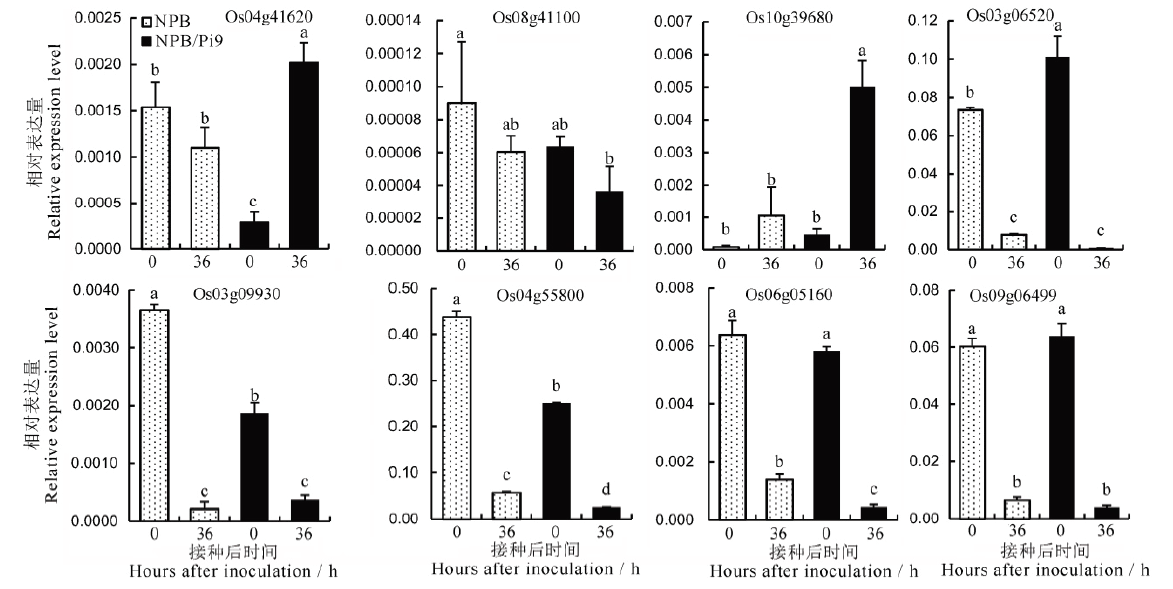
Fig. 5. qRT-PCR analysis of differentially expressed genes conferring chitinase and sulfate transporter after inoculation. Values are Mean±SD; Common lowercase letters indicate no significant difference at 0.05 level among treatments. The same as in Table 6.
| 基因 Gene | 12hexp-NPB/ 0hexp-NPB | 24hexp-NPB/ 0hexp-NPB | 36hexp-NPB/ 0hexp-NPB | 12hexp-Pi9/ 0hexp-Pi9 | 24hexp-Pi9/ 0hexp-Pi9 | 36hexp-Pi9/ 0hexp-Pi9 |
|---|---|---|---|---|---|---|
| Os10g39680 | 1.401 | 4.520 | 9.256 | 0.444 | 5.156 | 27.994 |
| Os04g41620 | 3.054 | 7.629 | 1.449 | 3.305 | 13.002 | 16.374 |
| Os08g41100 | 1.456 | 1.419 | 1.284 | 2.216 | 2.300 | 3.523 |
| Os03g06520 | 0.081 | 0.159 | 0.755 | 0.091 | 0.071 | 0.241 |
| Os03g09930 | 0.220 | 0.207 | 1.100 | 0.218 | 0.130 | 0.172 |
| Os04g55800 | 0.190 | 0.074 | 1.178 | 0.280 | 0.052 | 0.389 |
| Os06g05160 | 0.234 | 0.112 | 1.304 | 0.306 | 0.117 | 0.292 |
| Os09g06499 | 0.467 | 0.598 | 0.365 | 0.474 | 0.475 | 0.271 |
Table 5 Gene-chip detection data of rice chitinase and sulfate transporter genes in NPB/Pi9 and NPB.
| 基因 Gene | 12hexp-NPB/ 0hexp-NPB | 24hexp-NPB/ 0hexp-NPB | 36hexp-NPB/ 0hexp-NPB | 12hexp-Pi9/ 0hexp-Pi9 | 24hexp-Pi9/ 0hexp-Pi9 | 36hexp-Pi9/ 0hexp-Pi9 |
|---|---|---|---|---|---|---|
| Os10g39680 | 1.401 | 4.520 | 9.256 | 0.444 | 5.156 | 27.994 |
| Os04g41620 | 3.054 | 7.629 | 1.449 | 3.305 | 13.002 | 16.374 |
| Os08g41100 | 1.456 | 1.419 | 1.284 | 2.216 | 2.300 | 3.523 |
| Os03g06520 | 0.081 | 0.159 | 0.755 | 0.091 | 0.071 | 0.241 |
| Os03g09930 | 0.220 | 0.207 | 1.100 | 0.218 | 0.130 | 0.172 |
| Os04g55800 | 0.190 | 0.074 | 1.178 | 0.280 | 0.052 | 0.389 |
| Os06g05160 | 0.234 | 0.112 | 1.304 | 0.306 | 0.117 | 0.292 |
| Os09g06499 | 0.467 | 0.598 | 0.365 | 0.474 | 0.475 | 0.271 |
| 基因 Gene | 12hexp-NPB/ 0hexp-NPB | 24hexp-NPB/ 0hexp-NPB | 36hexp-NPB/ 0hexp-NPB | 12hexp-Pi9/ 0hexp-Pi9 | 24hexp-Pi9/ 0hexp-Pi9 | 36hexp-Pi9/ 0hexp-Pi9 |
|---|---|---|---|---|---|---|
| Os01g58280 | 0.783 | 3.735 | 22.434 | 1.527 | 15.248 | 177.553 |
| Os03g03550 | 0.339 | 0.112 | 34.030 | 0.119 | 0.081 | 0.142 |
| Os06g14190 | 0.330 | 0.656 | 9.012 | 0.183 | 0.395 | 0.496 |
| Os04g27670 | 3.134 | 54.153 | 15.309 | 5.077 | 112.337 | 115.703 |
| Os05g31040 | 4.042 | 24.376 | 59.293 | 1.669 | 1.520 | 0.398 |
| Os05g41290 | 0.867 | 3.768 | 2.319 | 4.637 | 7.419 | 12.060 |
| Os08g07340 | 13.957 | 10.544 | 7.274 | 0.559 | 0.395 | 0.345 |
| Os10g03850 | 8.021 | 2.133 | 0.796 | 13.489 | 7.518 | 6.698 |
Table 6 Gene-chip detection data of rice genes falling into different functional categories in NPB/Pi9 and NPB.
| 基因 Gene | 12hexp-NPB/ 0hexp-NPB | 24hexp-NPB/ 0hexp-NPB | 36hexp-NPB/ 0hexp-NPB | 12hexp-Pi9/ 0hexp-Pi9 | 24hexp-Pi9/ 0hexp-Pi9 | 36hexp-Pi9/ 0hexp-Pi9 |
|---|---|---|---|---|---|---|
| Os01g58280 | 0.783 | 3.735 | 22.434 | 1.527 | 15.248 | 177.553 |
| Os03g03550 | 0.339 | 0.112 | 34.030 | 0.119 | 0.081 | 0.142 |
| Os06g14190 | 0.330 | 0.656 | 9.012 | 0.183 | 0.395 | 0.496 |
| Os04g27670 | 3.134 | 54.153 | 15.309 | 5.077 | 112.337 | 115.703 |
| Os05g31040 | 4.042 | 24.376 | 59.293 | 1.669 | 1.520 | 0.398 |
| Os05g41290 | 0.867 | 3.768 | 2.319 | 4.637 | 7.419 | 12.060 |
| Os08g07340 | 13.957 | 10.544 | 7.274 | 0.559 | 0.395 | 0.345 |
| Os10g03850 | 8.021 | 2.133 | 0.796 | 13.489 | 7.518 | 6.698 |
| [1] | Deng Y, Zhai K, Xie Z, Yang D, Zhu X, Liu J, Wang X, Qin P, Yang Y, Zhang G, Li Q, Zhang J, Wu S, Milazzo J, Mao B, Wang E, Xie H, Tharreau D, He Z.Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance[J]. Science, 2017, 355(6328): 962-965. |
| [2] | Jain P, Singh P K, Kapoor R, Khanna A, Solanke A U, Krishnan S G, Singh A K, Sharma V, Sharma T R.Understanding host-pathogen interactions with expression profiling of NILs carrying rice-blast resistance Pi9 gene[J]. Frontiers of Plant Sciences, 2017, (8): 93-93. |
| [3] | Liu J, Hu Y, Ning Y, Jiang N, Wu J, Jeon J, Xiao Y, Liu X, Dai N, Wang G.Genetic variation and evolution of the Pi9 blast resistance locus in the AA genome Oryza species[J]. Journal of Plant Biology, 2011, 54(5): 294-302. |
| [4] | Wu J, Kou Y, Bao J, Li Y, Tang M, Zhu X, Ponaya A, Xiao G, Li J, Li C, Song M Y, Cumagun C J, Deng Q, Lu G, Jeon J S, Naqvi N I, Zhou B.Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9 that triggers Pi9-mediated blast resistance in rice[J]. New Phytologist, 2015, 206(4): 1463-1475. |
| [5] | Jones J D, Dang I J L. The plant immune system[J]. Nature, 2006, 444(7117): 323-329. |
| [6] | Liu W, Liu J, Triplett L, Leach J E, Wang G L.Novel insights into rice innate immunity against bacterial and fungal pathogens[J]. Phytopathology, 2014, 52(11): 213-241. |
| [7] | Kou Y, Wang S.Broad-spectrum and durability: Understanding of quantitative disease resistance[J]. Current Opinion in Plant Biology, 2010, 13: 181-185. |
| [8] | Wei T, Ou B, Li J, Zhao Y, Guo D, Zhu Y, Chen Z, Gu H, Li C, Qin G, Qu L J.Transcriptional profiling of rice early response to Magnaporthe oryzae identified OsWRKYs as important regulators in rice blast resistance[J]. PLoS One, 2013, 8(3): e59720. |
| [9] | Zhang Y, Zhao J, Li Y, Yuan Z, He H, Yang H, Qu H, Ma C, Qu S.Transcriptome analysis highlights defense and signaling pathways mediated by rice pi21 gene with partial resistance to Magnaporthe oryzae[J]. Frontiers of Plant Sciences, 2016, 7: 1834. |
| [10] | Li W, Liu Y, Wang J, He M, Zhou X, Yang C, Yuan C, Wang J, Chern M, Yin J, Chen W, Ma B, Wang Y, Qin P, Li S, Ronald P, Chen X.The durably resistant rice cultivar Digu activates defence gene expression before the full maturation of Magnaporthe oryzae appressorium[J]. Molecular Plant Pathology, 2015, 16: 973-986. |
| [11] | Kitony J K.水稻CO39品种及其5个近等基因系对稻瘟病感染应答的转录组分析[D]. 福州: 福建农林大学, 2016. |
| Kitony J K.Transcriptome analysis of rice cultivar CO39 and its five Near-Isogenic Lines in response to blast fungus infection[D]. Fuzhou: Fujian University of Agriculture and Forestry, 2016. (in Chinese with English abstract) | |
| [12] | 王玉, 杨雪, 杨蕊菁, 王玉霞, 杨飞霞, 夏鹏飞, 赵磊. 调控苯丙烷类生物合成的MYB类转录因子研究进展[J]. 安徽农业大学学报, 2019, 46(5): 859-864. |
| Wang Y, Yang X, Yang R Q, Wang Y X, Yang F X, Xia P F, Zhao L.Advances in research of MYB transcription factors in regulating phenylpropane biosynthesis[J]. Journal of Anhui Agricultural University, 2019, 46(5): 859-864. (in Chinese with English abstract) | |
| [13] | 崔慧萍, 周薇, 郭长虹. 植物过氧化物酶体在活性氧信号网络中的作用[J]. 中国生物化学与分子生物学报, 2017, 33(3): 220-226. (in Chinese with English abstract) |
| Cui H P, Zhou W, Guo C H.The role of plant peroxisomes in ROS signaling network[J]. Chinese Journal of Biochemistry and Molecular Biology, 2017, 33(3): 220-226. (in Chinese with English abstract) | |
| [14] | 杜欣谊. 苯丙氨酸解氨酶的研究进展[J]. 现代化农业, 2016(7): 24-26. |
| Du X Y.Research advances on phenylalanine ammonia-lyase gene[J]. Modernizing Agriculture, 2016(7): 24-26. (in Chinese with English abstract) | |
| [15] | 李魏, 谭晓风, 陈鸿鹏. 植物肉桂酰辅酶A还原酶基因的结构功能及应用潜力[J]. 经济林研究, 2009, 27(1): 7-12. |
| Li W, Tan X F, Chen H P.Structural function and application potential of cinnamoyl-CoA reductase gene in plant[J]. Non-wood Forest Research, 2009, 27(1): 7-12. (in Chinese with English abstract) | |
| [16] | Kasprzewska A.Plant chitinases-regulation and function[J]. Cellular & Molecular Biology Letters, 2003, 8(3): 809-824. |
| [17] | 许明辉, 李成云, 李进斌, 谭学林, 田颖川, 陈正华, 唐祚舜, 田文忠. 转几丁质酶-葡聚糖酶基因水稻稻瘟病抗谱分析[J]. 中国水稻科学, 2003, 17(4): 307-310. |
| Xu M H, Li C Y, Li J B, Tan X L, Tian Y C, Chen Z H, Tang Z S, Tian W Z.Analysis on resistant spectrum to rice blast in transgenic rice lines with chitinase gene and β-1, 3-glucanase gene[J]. Chinese Journal of Rice Science, 2003, 17(4): 307-310. (in Chinese with English abstract) | |
| [18] | Qu S, Liu G, Zhou B, Bellizzi M, Zeng L, Dai L, Han B, Wang G L.The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice[J]. Genetics, 2006, 172(3): 1901-1914. |
| [19] | Liu W, Liu J, Ning Y, Ding B, Wang X, Wang Z, Wang G.Recent progress in understanding PAMP- and effector-triggered immunity against the rice blast fungus Magnaporthe oryzae[J]. Molecular Plant, 2013, 6(3): 605-620. |
| [20] | Zipfel C.Pattern-recognition receptors in plant innate immunity[J]. Current Opinion Immunology, 2008, 20(1): 10-16. |
| [21] | Ramamoorthy R, Jiang S Y, Kumar N, Venkatesh P N, Ramachandran S.A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatment[J]. Plant Cell Physiology, 2008, 49(6): 865-879. |
| [22] | Peng X, Hu Y, Tang X, Zhou P, Deng X, Wang H, Guo Z.Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice[J]. Planta, 2012, 236(5): 1485-1498. |
| [23] | Pitzschke A, Schikora A, Hirt H.MAPK cascade signalling networks in plant defence[J]. Current Opinion in Plant Biology, 2009, 12(4): 421-426. |
| [1] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [2] | GAO Junru, QUAN Hongyu, YUAN Liuzhen, LI Qinying, QIAO Lei, LI Wenqiang. Map-based Cloning and Functional Analysis of a New Allele of D1, a Gene Controlling Plant Height in Rice (Oryza sativa L.) [J]. Chinese Journal OF Rice Science, 2024, 38(2): 140-149. |
| [3] | CHEN Mingliang, XIONG Wentao, SHEN Yumin, XIONG Huanjin, LUO Shiyou, WU Xiaoyan, HU Lanxiang, XIAO Yeqing. Genetic Dissection of Broad Spectrum Resistance of the Rice Maintainer Ganxiang B [J]. Chinese Journal OF Rice Science, 2023, 37(5): 470-477. |
| [4] | LI Gang, GAO Qingsong, LI Wei, ZHANG Wenxia, WANG Jian, CHEN Baoshan, WANG Di, GAO Hao, XU Weijun, CHEN Hongqi, JI Jianhui. Directed Knockout of SD1 Gene Improves Lodging Resistance and Blast Resistance of Rice [J]. Chinese Journal OF Rice Science, 2023, 37(4): 359-367. |
| [5] | FU Rongtao, WANG Jian, CHEN Cheng, ZHAO Liyu, CHEN Xuejuan, LU Daihua. Transcriptome Analysis of Young Rice Panicles in Early Response to Exposure to Mycotoxin of Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2022, 36(5): 447-458. |
| [6] | ZHOU Yonglin, SHEN Xiaolei, ZHOU Lishuai, LIN Qiaoxia, WANG Zhaolu, CHEN Jing, FENG Huijie, ZHANG Zhenwen, CHEN Xiaoting, LU Guodong. OsLOX10 Positively Regulates Defense Responses of Rice to Rice Blast and Bacterial Blight [J]. Chinese Journal OF Rice Science, 2022, 36(4): 348-356. |
| [7] | Huan CUI, Qiaoli GAO, Lixin LUO, Jing YANG, Chun CHEN, Tao GUO, Yongzhu LIU, Yongxiang HUANG, Hui WANG, Zhiqiang CHEN, Wuming XIAO. Transcriptome Analysis of Hormone Signal Transduction and Glutathione Metabolic Pathway in Rice Seeds at Germination Stage [J]. Chinese Journal OF Rice Science, 2021, 35(6): 554-564. |
| [8] | Yudong CAO, Xiangyi XIAO, Naizhong YE, Xiaowen DING, Xiaoxuan YI, Jinling LIU, Yinghui XIAO. Auxin Regulator OsGRF4 Simultaneously Regulates Rice Grain Shape and Blast Resistance [J]. Chinese Journal OF Rice Science, 2021, 35(6): 629-638. |
| [9] | Wei LIU, Zhanhua LU, Dongbai LU, Xiaofei WANG, Shiguang WANG, Jia XUE, Xiuying HE. Location and Candidate Gene Analysis of Rice Clustered Spikelets Gene OsCL6 [J]. Chinese Journal OF Rice Science, 2021, 35(3): 238-248. |
| [10] | Huajun ZHU, Hu ZHOU, Zuohua REN, Erming LIU. Extracellular Antimicrobial Substances Produced by Bacillus subtilis JN005 and Its Control Efficacy on Rice Leaf Blast [J]. Chinese Journal OF Rice Science, 2020, 34(5): 470-478. |
| [11] | Jinlu LI, Hui ZHANG, Zeyu JIAO, Jianyu LIU, Guangyu HAN, Xiaoxuan ZHUO, Qiong LUO. Identification of Blast Disease Resistance-related Genes by Genomic Sequence Comparison of Rice Variety Ziyu 44 and Jiangnanxiangnuo [J]. Chinese Journal OF Rice Science, 2020, 34(1): 8-16. |
| [12] | Tao CHEN, Xuchao SUN, Shanlei ZHANG, Wenhua LIANG, Lihui ZHOU, Qingyong ZHAO, Shu YAO, Ling ZHAO, Chunfang ZHAO, Zhen ZHU, Yadong ZHANG, Cailin WANG. Development and Verification of Specific Molecular Markers for Pigm Gene Associated with Broad-spectrum Resistance to Rice Blast [J]. Chinese Journal OF Rice Science, 2020, 34(1): 28-36. |
| [13] | Ni CAO, Yuan CHEN, Zhijuan JI, Yuxiang ZENG, Changdeng YANG, Yan LIANG. Recent Progress in Molecular Mechanism of Rice Blast Resistance [J]. Chinese Journal OF Rice Science, 2019, 33(6): 489-498. |
| [14] | Yan LIU, Baoxiang WANG, Bo YANG, Tingmu CHEN, Yungao XING, Zhiguang SUN, Bo XU, Ming CHI, Baiguan LU, Jian LI, Jinbo LIU, Zhaowei FANG, Derong QIN, Dayong XU. Genetic Analysis of Rice Neck Blast Resistance in Huang-Huai-Hai Region [J]. Chinese Journal OF Rice Science, 2019, 33(4): 377-382. |
| [15] | Zhengyan PAN, Fulin QIU, Guilan LÜ, Xiufang MA, Wenqi SHANG, Yuanjun BAI, Zhengjin XU. Analysis of Rice Blast Resistance Genes in japonica Rice Varieties in Liaoning Province [J]. Chinese Journal OF Rice Science, 2019, 33(3): 241-248. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||