
Chinese Journal OF Rice Science ›› 2018, Vol. 32 ›› Issue (6): 529-537.DOI: 10.16819/j.1001-7216.2018.8001
• Orginal Article • Previous Articles Next Articles
Minjuan ZHANG1,2, Shuaijun LI1,2, Qiongqiong CHEN1, Xiuqing JING1, Kunming CHEN1, Chunhai SHI3,*( ), Wenqiang LI1,*(
), Wenqiang LI1,*( )
)
Received:2018-01-07
Revised:2018-02-10
Online:2018-11-27
Published:2018-11-10
Contact:
Chunhai SHI, Wenqiang LI
张敏娟1,2, 李帅军1,2, 陈琼琼1, 景秀清1, 陈坤明1, 石春海3,*( ), 李文强1,*(
), 李文强1,*( )
)
通讯作者:
石春海,李文强
CLC Number:
Minjuan ZHANG, Shuaijun LI, Qiongqiong CHEN, Xiuqing JING, Kunming CHEN, Chunhai SHI, Wenqiang LI. Genetic Mapping and Proteomic Analysis of the Dwarf and Low-tillering Mutant dlt3 in Rice[J]. Chinese Journal OF Rice Science, 2018, 32(6): 529-537.
张敏娟, 李帅军, 陈琼琼, 景秀清, 陈坤明, 石春海, 李文强. 水稻矮化少蘖突变体dlt3的基因定位和蛋白质组学分析[J]. 中国水稻科学, 2018, 32(6): 529-537.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2018.8001
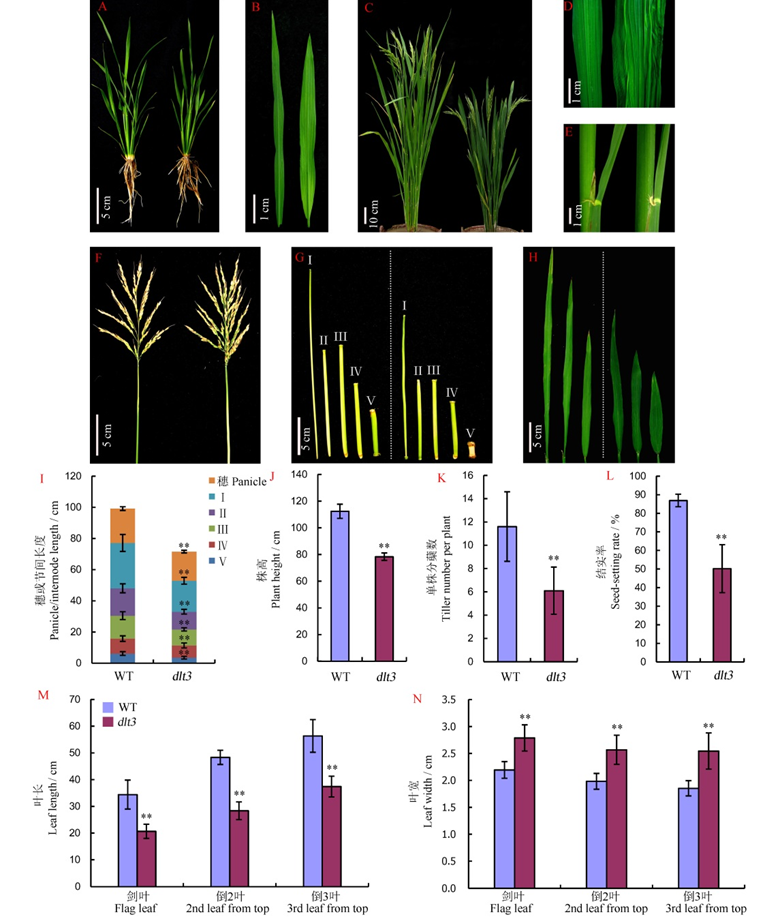
Fig. 1. Phenotypic characterization of WT and dlt3 mutant.^ A and B, Plant and leaf blade at the tillering stage. C to H, Morphology of mature plant, middle portion of blade, lamina joint, panicle, internodes and top three leaf blades. The left represents wild type and the right the dlt3 mutant. Mean±SD, n=12. **Difference between mutant and WT was significant at 0.01 level.
| 标记名称 Marker name | 正向引物序列(5′-3′) Forward primer sequence | 反向引物序列(5′-3′) Reverse primer sequence | 染色体 Chromosome | 物理位置 Physical position/kb |
|---|---|---|---|---|
| RM1163 | TGGACGCGGATAGGAGGAGACG | TCCTCCGCAAGGTCGGTTTCC | 6 | 4021 |
| RM3408 | AGTGAATCCCATCAGATCACTCC | CATATCAAATCGCCGAGAAGG | 6 | 4608 |
| RM19556 | TTCTGGCCTATGAGGGATGATCG | ACCAAATATCAAGCCTGCACAGC | 6 | 5249 |
| RM19570 | CCCAGATATTCTGTGTGATCATGAGG | GAGTGAATGTGAGCCGTCTATTGG | 6 | 5422 |
| RM2615 | ATCTCGTTCATACTGCTTGACC | GACTGGTTTCCTTCATGTTACC | 6 | 5970 |
| RM276 | GTCCTCCATCGAGCAGTATCAGC | CTAGCAAGACATGGACCTCAACG | 6 | 6241 |
| RM19638 | CCACACTGTACCGGTCGAAGACG | CTCTACAACTTGCAGCCCTGTCAGC | 6 | 6387 |
| R6M14 | AAATGTCCATGTGTTTGCTTC | CATGTGTGGAATGTGGTTG | 6 | 7710 |
| RM527 | CGGTTTGTACGTAAGTAGCATCAGG | TCCAATGCCAACAGCTATACTCG | 6 | 9874 |
Table 1 Molecular markers and its sequences used for genetic mapping of dlt3 in this study.
| 标记名称 Marker name | 正向引物序列(5′-3′) Forward primer sequence | 反向引物序列(5′-3′) Reverse primer sequence | 染色体 Chromosome | 物理位置 Physical position/kb |
|---|---|---|---|---|
| RM1163 | TGGACGCGGATAGGAGGAGACG | TCCTCCGCAAGGTCGGTTTCC | 6 | 4021 |
| RM3408 | AGTGAATCCCATCAGATCACTCC | CATATCAAATCGCCGAGAAGG | 6 | 4608 |
| RM19556 | TTCTGGCCTATGAGGGATGATCG | ACCAAATATCAAGCCTGCACAGC | 6 | 5249 |
| RM19570 | CCCAGATATTCTGTGTGATCATGAGG | GAGTGAATGTGAGCCGTCTATTGG | 6 | 5422 |
| RM2615 | ATCTCGTTCATACTGCTTGACC | GACTGGTTTCCTTCATGTTACC | 6 | 5970 |
| RM276 | GTCCTCCATCGAGCAGTATCAGC | CTAGCAAGACATGGACCTCAACG | 6 | 6241 |
| RM19638 | CCACACTGTACCGGTCGAAGACG | CTCTACAACTTGCAGCCCTGTCAGC | 6 | 6387 |
| R6M14 | AAATGTCCATGTGTTTGCTTC | CATGTGTGGAATGTGGTTG | 6 | 7710 |
| RM527 | CGGTTTGTACGTAAGTAGCATCAGG | TCCAATGCCAACAGCTATACTCG | 6 | 9874 |
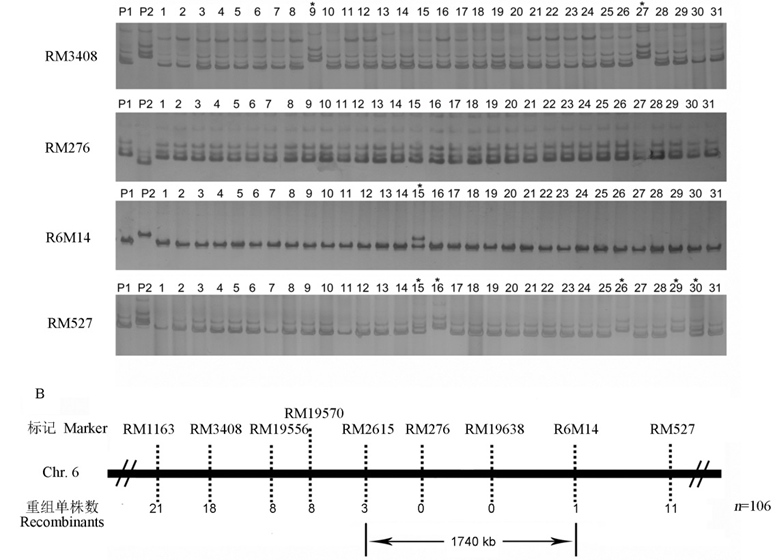
Fig. 3. Genetic linkage analysis and molecular mapping of dlt3 gene.^ A, Genotyping of F2 mutant plants by dlt3-linked molecular markers RM3408, RM276, R6M14 and RM527; B, Genetic mapping of dlt3 gene by the molecular markers. *Recombinant.
| 蛋白登录号 Protein accession | 水稻基因号 RAP_locus | 蛋白质功能描述 Protein description | 表达量变化 Ratio (dlt3/WT) | P值 P-value | 上(下)调 Regulation |
|---|---|---|---|---|---|
| 油菜素内酯相关蛋白 Brassinosteroid related proteins | |||||
| Q6Z730 | Os02g0122300 | Translation initiation factor eIF3 subunit domain containing protein, Brassinosteroid receptor kinase (BRI1)-interacting protein 121, BIP121 | 0.651 | 1.08×10-2 | Down |
| Q0JBZ3 | Os04g0501600 | AT hook, DNA-binding, conserved site domain containing protein, brassinosteroid receptor kinase (BRI1)-interacting protein 106, BIP106 | 1.372 | 9.26×10-3 | Up |
| Q6K624 | Os02g0612800 | AT hook, DNA-binding, conserved site domain containing protein, brassinosteroid receptor kinase (BRI1)-interacting protein 135, BIP135 | 1.374 | 4.11×10-2 | Up |
| Q0J5J5 | Os08g0430500 | 14-3-3 protein, Florigen receptor, G-box factor 14-3-3c protein, OsGF14c | 3.355 | 3.15×10-3 | Up |
| 生长素相关蛋白 Auxin related proteins | |||||
| Q0DSY8 | Os03g0280000 | Similar to MDR-like ABC transporter, P-Glycoprotein 13, OsABCB13, OsPGP13 | 1.329 | 2.71×10-2 | Up |
| Q0IR61 | Os11g0673200 | Similar to Auxin-induced beta-glucosidase | 1.828 | 1.65×10-2 | Up |
| Q84QW4 | Os08g0524400 | Protein of unknown function DUF568, DOMON-like domain containing protein, auxin-responsive family protein AIR12 | 1.857 | 4.48×10-2 | Up |
| 激酶和磷酸酯酶相关蛋白 Kinase and phosphatase related proteins | |||||
| Q0D6J6 | Os07g0472400 | Serine/threonine protein kinase-related domain containing protein | 0.548 | 1.01×10-2 | Down |
| Q69NF8 | Os09g0529900 | Pyruvate/Phosphoenolpyruvate kinase, catalytic core domain containing protein | 0.716 | 8.12E-03 | Down |
| Q5Z8F2 | Os01g0337600 | Suppressor of Mek, PH domain-like protein | 0.697 | 1.38×10-2 | Down |
| Q0D4F7 | Os07g0628700 | Similar to Receptor protein kinase | 1.328 | 1.51×10-2 | Up |
| Q5QLG3 | Os01g0682500 | Similar to Protein Kinase C630.09c | 1.431 | 1.86×10-3 | Up |
| Q2R2T6 | Os11g0549615 | Ser/Thr protein phosphatase family protein, purple acid phosphatase 3a | 1.889 | 2.72×10-2 | Up |
| Q0IP84 | Os12g0236400 | Adenylate kinase A (EC 2.7.4.3) (ATP-AMP transphosphorylase) | 1.471 | 1.44×10-2 | Up |
| Q60DT7 | Os05g0190500 | Similar to acid phosphatase, HAD superfamily phosphatase, VEGETATIVE STORAGE PROTEIN 2, OsVSP2 | 1.550 | 3.50×10-2 | Up |
| Q6AVA8 | Os05g0405000 | floury endosperm 4, pyruvate orthophospate dikinase, OsPBDK | 1.641 | 7.29×10-4 | Up |
| Q6H444 | Os09g0279400 | Rhodanese-like domain containing protein, Rhodanese / Cell cycle control phosphatase superfamily protein | 2.096 | 8.54×10-3 | Up |
| Q0IPL3 | Os12g0189300 | Pyruvate/Phosphoenolpyruvate kinase, catalytic core domain containing protein | 2.248 | 6.06×10-4 | Up |
| Q6KA61 | Os02g0285800 | Similar to GTP-binding protein typA (Tyrosine phosphorylated protein A) | 3.491 | 3.91×10-4 | Up |
| Q7XQZ5 | Os04g0661300 | ATP/GTP binding protein | 1.356 | 1.09×10-2 | Up |
| Q0JQ62 | Os01g0179700 | Similar to GTP-binding protein YPTM2, ras-related protein, OsRab1 | 1.302 | 2.27×10-2 | Up |
| Q0DH67 | Os05g0489600 | ADP-ribosylation factor, Ras-related small GTP-binding family protein | 1.608 | 3.99×10-3 | Up |
| Q84T71 | Os03g0854100 | Similar to ARF GAP-like zinc finger-containing protein ZIGA2, GTPase-activating protein | 1.322 | 3.38×10-3 | Up |
| 钙结合蛋白 Ca2+ binding related proteins | |||||
| Q6F334 | Os05g0491000 | EF-Hand type domain containing protein, calmodulin-like protein 9, OsCML9 | 0.621 | 1.71×10-3 | Down |
| Q84VG0 | Os08g0117400 | Calmodulin-related calcium sensor protein, Ser/Thr kinase/calmodulin, OsCML7, OsSTKC | 0.732 | 7.55×10-3 | Down |
| Q0DFA8 | Os06g0104400 | IQ calmodulin-binding motif family protein | 1.458 | 1.17×10-4 | Up |
| Q0D7H6 | Os07g0246200 | Similar to calreticulin, CRT | 1.325 | 6.48×10-6 | Up |
| 锌指结构域蛋白 Zinc finger containing proteins | |||||
| Q6K977 | Os02g0831100 | Zinc finger CCCH domain-containing protein 19 | 0.400 | 4.02×10-3 | Down |
| Q2R4J4 | Os11g0472000 | Zinc finger CCCH-type domain containing protein 63 | 0.436 | 2.02×10-3 | Down |
| Q6Z6E6 | Os02g0203700 | Zinc finger family protein, SRZ1 | 0.563 | 5.50×10-3 | Down |
| Q5NAV3 | Os01g0257400 | Zinc finger CCCH-type domain containing protein, OsC3H5 | 0.638 | 1.30×10-2 | Down |
| Q6YUR8 | Os02g0121100 | Zinc finger CCHC-type domain containing protein, cold shock domain protein 1, CSD protein 1 | 0.679 | 8.90×10-3 | Down |
| Q6K4N0 | Os02g0789400 | Similar to 9G8-like SR protein, RNA recognition motif and CCHC-type zinc finger domains containing protein | 1.332 | 4.31×10-2 | Up |
| Q75LJ7 | Os03g0836200 | Similar to RNA-binding protein RZ-1, zinc finger-containing glycine-rich RNA-binding protein 1 | 1.374 | 2.39×10-3 | Up |
| Q6YTY3 | Os07g0608400 | PHD/F-BOX containing protein, PHD zinc finger protein ALFIN-LIKE 9 | 1.313 | 1.73×10-2 | Up |
| 水稻株型调控相关蛋白 Regulation of plant architecture and growth related proteins | |||||
| Q6EU14 | Os02g0672200 | Similar to AGO1 homologous protein, OsAGO1a | 0.471 | 1.96×10-3 | Down |
| Q2QN08 | Os12g0583500 | Broad Complex BTB domain with non-phototropic hypocotyl 3 NPH3 domain, BTB domain containing protein | 0.375 | 9.84×10-3 | Down |
| Q0J0F4 | Os09g0513000 | Similar to TGB12K interacting protein 3, ankyrin repeat domain containing protein, XA21 binding protein 25 | 0.712 | 3.06×10-2 | Down |
| Q10L71 | Os03g0356700 | Villin/gelsolin superfamily protein, Actin binding protein, Regulation of plant architecture, VLN2 | 1.350 | 2.66×10-2 | Up |
| Q0ITK1 | Os11g0247300 | Tubulin/FtsZ domain containing protein, Cell elongation and division, TWISTED DWARF 1, Tubulin alpha-2, small and round seed 5, TID1, TubA2, SRS5 | 2.936 | 3.45×10-3 | Up |
| Q0DH37 | Os05g0494000 | Cytochrome P450 | 1.507 | 4.15×10-3 | Up |
| Q2R176 | Os11g0615200 | SSXT family protein, GRF-interacting factor 2, protein binding/transcription coactivator | 1.428 | 2.04×10-2 | Up |
| Q7XHW8 | Os07g0681000 | Zinc-binding domain of translation initiation factor 2 beta | 1.429 | 4.84×10-4 | Up |
| Q0JAK9 | Os04g0592400 | Protein transport protein-related, t-snare domain containing protein | 1.574 | 9.88×10-4 | Up |
| Q0JP92 | Os01g0235400 | Similar to Importin-alpha re-exporter, Cellular apoptosis susceptibility protein homolog | 2.102 | 1.54×10-2 | Up |
Table 2 Selected differentially expressed proteins identified by iTRAQ-based quantitative proteomic analysis.
| 蛋白登录号 Protein accession | 水稻基因号 RAP_locus | 蛋白质功能描述 Protein description | 表达量变化 Ratio (dlt3/WT) | P值 P-value | 上(下)调 Regulation |
|---|---|---|---|---|---|
| 油菜素内酯相关蛋白 Brassinosteroid related proteins | |||||
| Q6Z730 | Os02g0122300 | Translation initiation factor eIF3 subunit domain containing protein, Brassinosteroid receptor kinase (BRI1)-interacting protein 121, BIP121 | 0.651 | 1.08×10-2 | Down |
| Q0JBZ3 | Os04g0501600 | AT hook, DNA-binding, conserved site domain containing protein, brassinosteroid receptor kinase (BRI1)-interacting protein 106, BIP106 | 1.372 | 9.26×10-3 | Up |
| Q6K624 | Os02g0612800 | AT hook, DNA-binding, conserved site domain containing protein, brassinosteroid receptor kinase (BRI1)-interacting protein 135, BIP135 | 1.374 | 4.11×10-2 | Up |
| Q0J5J5 | Os08g0430500 | 14-3-3 protein, Florigen receptor, G-box factor 14-3-3c protein, OsGF14c | 3.355 | 3.15×10-3 | Up |
| 生长素相关蛋白 Auxin related proteins | |||||
| Q0DSY8 | Os03g0280000 | Similar to MDR-like ABC transporter, P-Glycoprotein 13, OsABCB13, OsPGP13 | 1.329 | 2.71×10-2 | Up |
| Q0IR61 | Os11g0673200 | Similar to Auxin-induced beta-glucosidase | 1.828 | 1.65×10-2 | Up |
| Q84QW4 | Os08g0524400 | Protein of unknown function DUF568, DOMON-like domain containing protein, auxin-responsive family protein AIR12 | 1.857 | 4.48×10-2 | Up |
| 激酶和磷酸酯酶相关蛋白 Kinase and phosphatase related proteins | |||||
| Q0D6J6 | Os07g0472400 | Serine/threonine protein kinase-related domain containing protein | 0.548 | 1.01×10-2 | Down |
| Q69NF8 | Os09g0529900 | Pyruvate/Phosphoenolpyruvate kinase, catalytic core domain containing protein | 0.716 | 8.12E-03 | Down |
| Q5Z8F2 | Os01g0337600 | Suppressor of Mek, PH domain-like protein | 0.697 | 1.38×10-2 | Down |
| Q0D4F7 | Os07g0628700 | Similar to Receptor protein kinase | 1.328 | 1.51×10-2 | Up |
| Q5QLG3 | Os01g0682500 | Similar to Protein Kinase C630.09c | 1.431 | 1.86×10-3 | Up |
| Q2R2T6 | Os11g0549615 | Ser/Thr protein phosphatase family protein, purple acid phosphatase 3a | 1.889 | 2.72×10-2 | Up |
| Q0IP84 | Os12g0236400 | Adenylate kinase A (EC 2.7.4.3) (ATP-AMP transphosphorylase) | 1.471 | 1.44×10-2 | Up |
| Q60DT7 | Os05g0190500 | Similar to acid phosphatase, HAD superfamily phosphatase, VEGETATIVE STORAGE PROTEIN 2, OsVSP2 | 1.550 | 3.50×10-2 | Up |
| Q6AVA8 | Os05g0405000 | floury endosperm 4, pyruvate orthophospate dikinase, OsPBDK | 1.641 | 7.29×10-4 | Up |
| Q6H444 | Os09g0279400 | Rhodanese-like domain containing protein, Rhodanese / Cell cycle control phosphatase superfamily protein | 2.096 | 8.54×10-3 | Up |
| Q0IPL3 | Os12g0189300 | Pyruvate/Phosphoenolpyruvate kinase, catalytic core domain containing protein | 2.248 | 6.06×10-4 | Up |
| Q6KA61 | Os02g0285800 | Similar to GTP-binding protein typA (Tyrosine phosphorylated protein A) | 3.491 | 3.91×10-4 | Up |
| Q7XQZ5 | Os04g0661300 | ATP/GTP binding protein | 1.356 | 1.09×10-2 | Up |
| Q0JQ62 | Os01g0179700 | Similar to GTP-binding protein YPTM2, ras-related protein, OsRab1 | 1.302 | 2.27×10-2 | Up |
| Q0DH67 | Os05g0489600 | ADP-ribosylation factor, Ras-related small GTP-binding family protein | 1.608 | 3.99×10-3 | Up |
| Q84T71 | Os03g0854100 | Similar to ARF GAP-like zinc finger-containing protein ZIGA2, GTPase-activating protein | 1.322 | 3.38×10-3 | Up |
| 钙结合蛋白 Ca2+ binding related proteins | |||||
| Q6F334 | Os05g0491000 | EF-Hand type domain containing protein, calmodulin-like protein 9, OsCML9 | 0.621 | 1.71×10-3 | Down |
| Q84VG0 | Os08g0117400 | Calmodulin-related calcium sensor protein, Ser/Thr kinase/calmodulin, OsCML7, OsSTKC | 0.732 | 7.55×10-3 | Down |
| Q0DFA8 | Os06g0104400 | IQ calmodulin-binding motif family protein | 1.458 | 1.17×10-4 | Up |
| Q0D7H6 | Os07g0246200 | Similar to calreticulin, CRT | 1.325 | 6.48×10-6 | Up |
| 锌指结构域蛋白 Zinc finger containing proteins | |||||
| Q6K977 | Os02g0831100 | Zinc finger CCCH domain-containing protein 19 | 0.400 | 4.02×10-3 | Down |
| Q2R4J4 | Os11g0472000 | Zinc finger CCCH-type domain containing protein 63 | 0.436 | 2.02×10-3 | Down |
| Q6Z6E6 | Os02g0203700 | Zinc finger family protein, SRZ1 | 0.563 | 5.50×10-3 | Down |
| Q5NAV3 | Os01g0257400 | Zinc finger CCCH-type domain containing protein, OsC3H5 | 0.638 | 1.30×10-2 | Down |
| Q6YUR8 | Os02g0121100 | Zinc finger CCHC-type domain containing protein, cold shock domain protein 1, CSD protein 1 | 0.679 | 8.90×10-3 | Down |
| Q6K4N0 | Os02g0789400 | Similar to 9G8-like SR protein, RNA recognition motif and CCHC-type zinc finger domains containing protein | 1.332 | 4.31×10-2 | Up |
| Q75LJ7 | Os03g0836200 | Similar to RNA-binding protein RZ-1, zinc finger-containing glycine-rich RNA-binding protein 1 | 1.374 | 2.39×10-3 | Up |
| Q6YTY3 | Os07g0608400 | PHD/F-BOX containing protein, PHD zinc finger protein ALFIN-LIKE 9 | 1.313 | 1.73×10-2 | Up |
| 水稻株型调控相关蛋白 Regulation of plant architecture and growth related proteins | |||||
| Q6EU14 | Os02g0672200 | Similar to AGO1 homologous protein, OsAGO1a | 0.471 | 1.96×10-3 | Down |
| Q2QN08 | Os12g0583500 | Broad Complex BTB domain with non-phototropic hypocotyl 3 NPH3 domain, BTB domain containing protein | 0.375 | 9.84×10-3 | Down |
| Q0J0F4 | Os09g0513000 | Similar to TGB12K interacting protein 3, ankyrin repeat domain containing protein, XA21 binding protein 25 | 0.712 | 3.06×10-2 | Down |
| Q10L71 | Os03g0356700 | Villin/gelsolin superfamily protein, Actin binding protein, Regulation of plant architecture, VLN2 | 1.350 | 2.66×10-2 | Up |
| Q0ITK1 | Os11g0247300 | Tubulin/FtsZ domain containing protein, Cell elongation and division, TWISTED DWARF 1, Tubulin alpha-2, small and round seed 5, TID1, TubA2, SRS5 | 2.936 | 3.45×10-3 | Up |
| Q0DH37 | Os05g0494000 | Cytochrome P450 | 1.507 | 4.15×10-3 | Up |
| Q2R176 | Os11g0615200 | SSXT family protein, GRF-interacting factor 2, protein binding/transcription coactivator | 1.428 | 2.04×10-2 | Up |
| Q7XHW8 | Os07g0681000 | Zinc-binding domain of translation initiation factor 2 beta | 1.429 | 4.84×10-4 | Up |
| Q0JAK9 | Os04g0592400 | Protein transport protein-related, t-snare domain containing protein | 1.574 | 9.88×10-4 | Up |
| Q0JP92 | Os01g0235400 | Similar to Importin-alpha re-exporter, Cellular apoptosis susceptibility protein homolog | 2.102 | 1.54×10-2 | Up |
| [1] | Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush G S, Kitano H, Matsuoka M.Green revolution: A mutant gibberellin-synthesis gene in rice.Nature, 2002, 416(6882): 701-702. |
| [2] | Hedden P.The genes of the green revolution.Trends Genet, 2003, 19(1): 5-9. |
| [3] | Smith S M, Li C, Li J.1-Hormone function in plants.Horm Metabol Signal Plants, 2017: 1-38. |
| [4] | Santner A, Calderon-Villalobos L I A,Estelle M. Plant hormones are versatile chemical regulators of plant growth.Nat Chem Biol, 2009, 5(5): 301-307. |
| [5] | Hedden P, Phillips A L.Gibberellin metabolism: New insights revealed by the genes.Trends Plant Sci, 2000, 5(12): 523-530. |
| [6] | Clouse S D, Sasse J M.Brassinosteroids: Essential regulators of plant growth and development.Annu Rev Plant Physiol Plant Mol Biol, 1998, 49(49): 427-451. |
| [7] | Lanahan M B, Ho T H (1988) Slender barley: A constitutive gibberellin-response mutant.Planta, 1988, 175(1): 107-114. |
| [8] | Tong H N, Jin Y, Liu W B, Li F, Fang J, Yin Y H, Qian Q, Zhu L H, Chu C C.DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J, 2009, 58(5): 803-816. |
| [9] | Murray M G, Thompson W F.Rapid isolation of high molecular weight plant DNA.Nucl Acids Res, 1980, 8(19): 4321-4325. |
| [10] | Wang Z Q, Xu X Y, Gong Q Q, Xie C, Fan W, Yang J L, Lin Q S, Zheng S J.Root proteome of rice studied by iTRAQ provides integrated insight into aluminum stress tolerance mechanisms in plants.J Proteomics, 2014, 98(4): 189-205. |
| [11] | 李磊, 薛芗, 左示敏, 陈宗祥, 张亚芳, 李前前, 朱俊凯, 马玉银, 潘学彪, 潘存红. 抑制OsAGO1a基因的表达导致水稻叶片近轴面卷曲. 中国水稻科学, 2013, 27(3): 223-230. |
| Li L, Xue X, Zuo S M, Chen Z X, Zhang Y F, Li Q Q, Zhu J K, Ma Y Y, Pan X B, Pan C H.Suppressed expression ofOsAGO1a leads to adaxial leaf rolling in rice. Chin J Rice Sci, 2013, 27(3): 223-230. | |
| [12] | Wu S Y, Xie Y R, Zhang J J, Ren Y L, Zhang X, Wang J L, Guo X P, Wu F Q, Sheng P K, Wang J, Wu C, Wang H, Huang S, Wan [J]. VLN2 regulates plant architecture by affecting microfilament dynamics and polar auxin transport in rice. Plant Cell, 2015, 27(10): 2829-2845. |
| [13] | Sunohara H, Kawai T, Shimizu-Sato S, Sato Y, Sato K, Kitano H.A dominant mutation of TWISTED DWARF 1 encoding an alpha-tubulin protein causes severe dwarfism and right helical growth in rice. Genes Genet Syst, 2009, 84(3): 209-218. |
| [14] | Li W, Wu J, Weng S, Zhang Y, Zhang D, Shi C.Identification and characterization of dwarf 62, a loss-of-function mutation in DLT/OsGRAS-32 affecting gibberellin metabolism in rice. Planta, 2010, 232(6): 1383-1396. |
| [15] | Tong H N, Liu L C, Jin Y, Du L, Yin Y H, Qian Q, Zhu L H, Chu C C.DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell, 2012, 24(6): 2562-2577. |
| [16] | Hirano K, Yoshida H, Aya K, Kawamura M, Hayashi M, Hobo T, Sato-Izawa K, Kitano H, Ueguchi-Tanaka M, Matsuoka M.SMALL ORGAN SIZE 1 and SMALL ORGAN SIZE 2/DWARF AND LOW-TILLERING form a complex to integrate auxin and brassinosteroid signaling in rice. Mol Plant, 2017, 10(4): 590-604. |
| [17] | Sun L, Li X, Fu Y, Zhu Z, Tan L, Liu F, Sun X, Sun X, Sun C.GS6, a member of the GRAS gene family, negatively regulates grain size in rice. J Integr Plant Biol, 2013, 55(10): 938-949. |
| [18] | Li D, Wang L, Wang M, Xu Y Y, Luo W, Liu Y J, Xu Z H, Li J, Chong K.EngineeringOsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J, 2009, 7(8): 791-806. |
| [19] | Tanaka A, Nakagawa H, Tomita C, Shimatani Z, Ohtake M, Nomura T, Jiang C, Dubouzet J G, Kikuchi S, Sekimoto H, Yokota T, Asami T, Kamakura T, Mori M.BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol, 2009, 151(2): 669-680. |
| [20] | Sakamoto T, Morinaka Y, Inukai Y, Kitano H, Fujioka S.Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J, 2013, 73(4): 676-688. |
| [21] | Purwestri Y A, Ogaki Y, Tamaki S, Tsuji H, Shimamoto K.The 14-3-3 protein GF14c acts as a negative regulator of flowering in rice by interacting with the florigen Hd3a.Plant Cell Physiol, 2009, 50(3): 429-438. |
| [22] | Bai M Y, Zhang L Y, Gampala S S, Zhu S W, Song W Y, Chong K, Wang Z Y.Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice.Proc Natl Acad Sci USA, 2007, 104(34): 13839-13844. |
| [23] | Boonburapong B, Buaboocha T.Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. .BMC Plant Biol, 2007, 7(1): 4. |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [12] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [13] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [14] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| [15] | WEI Qianqian, WANG Yulei, KONG Haimin, XU Qingshan, YAN Yulian, PAN Lin, CHI Chunxin, KONG Yali, TIAN Wenhao, ZHU Lianfeng, CAO Xiaochuang, ZHANG Junhua, ZHU Chunqun. Mechanism of Hydrogen Sulfide, a Signaling Molecule Involved in Reducing the Inhibitory Effect of Aluminum Toxicity on Rice Growth Together with Sulfur Fertilizer [J]. Chinese Journal OF Rice Science, 2024, 38(3): 290-302. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||