
Chinese Journal OF Rice Science ›› 2018, Vol. 32 ›› Issue (5): 415-426.DOI: 10.16819/j.1001-7216.2018.8003
• 研究论文 • Next Articles
Jingfang LI1, Yunlu TIAN1, Xi LIU1, Shijia LIU1, Liangming CHEN1, Ling JIANG1, Wenwei ZHANG1, Dayong XU2, Yihua WANG1,*( ), Jianmin WAN1
), Jianmin WAN1
Received:2018-01-15
Revised:2018-03-17
Online:2018-09-10
Published:2018-09-10
Contact:
Yihua WANG
李景芳1, 田云录1, 刘喜1, 刘世家1, 陈亮明1, 江玲1, 张文伟1, 徐大勇2, 王益华1,*( ), 万建民1
), 万建民1
通讯作者:
王益华
基金资助:CLC Number:
Jingfang LI, Yunlu TIAN, Xi LIU, Shijia LIU, Liangming CHEN, Ling JIANG, Wenwei ZHANG, Dayong XU, Yihua WANG, Jianmin WAN. The Guanylate Kinase OsGK1 is Essential for Seed Development in Rice[J]. Chinese Journal OF Rice Science, 2018, 32(5): 415-426.
李景芳, 田云录, 刘喜, 刘世家, 陈亮明, 江玲, 张文伟, 徐大勇, 王益华, 万建民. 鸟苷酸激酶OsGK1对水稻种子发育至关重要[J]. 中国水稻科学, 2018, 32(5): 415-426.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.16819/j.1001-7216.2018.8003
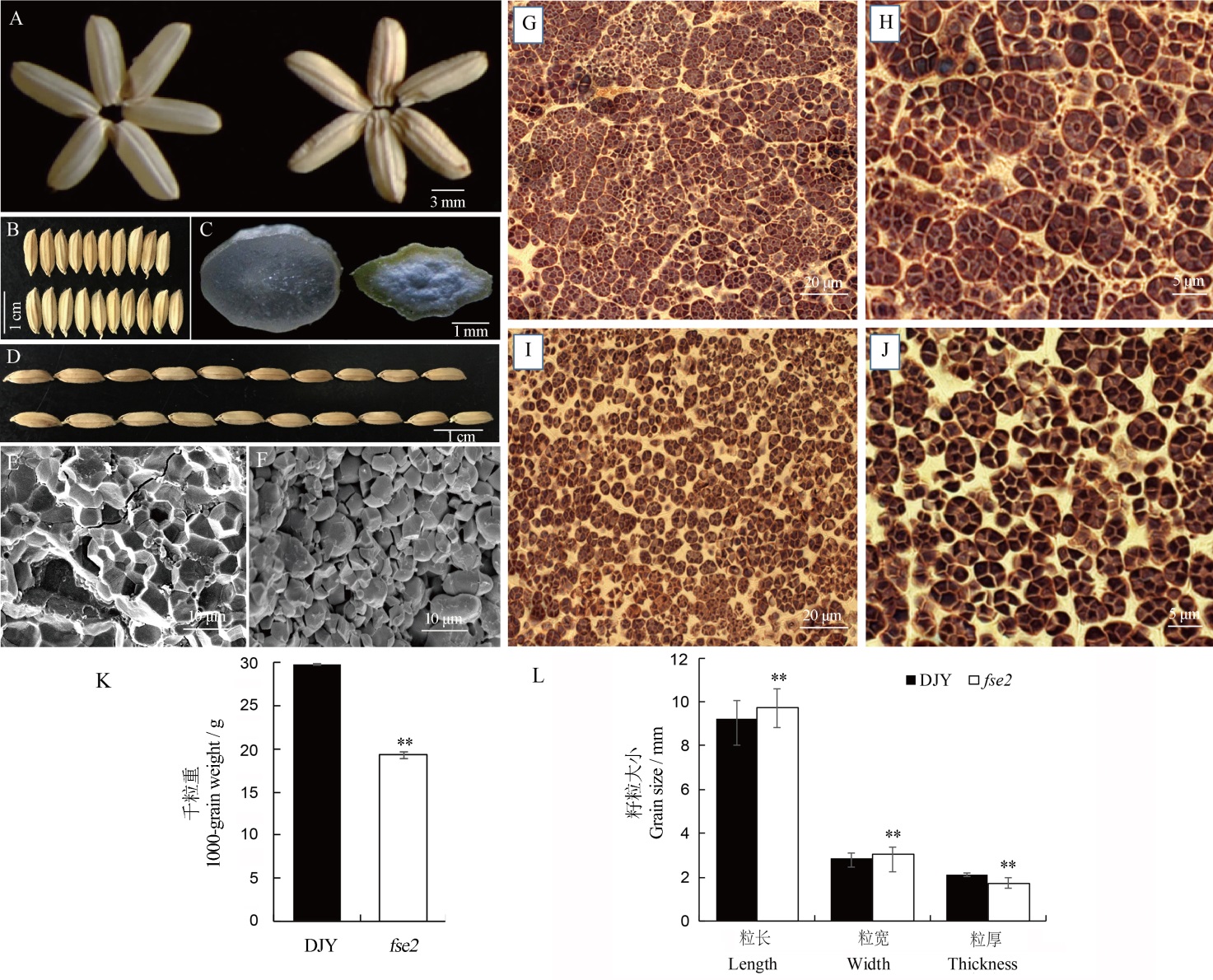
Fig. 1. Phenotypic comparison of mature seeds of wild type and fse2 mutant. A, B, D, Mature seeds of Dianjingyou 1(DJY) and fse2. A, DJY (left), fse2 (right). B, D, DJY (upper), fse2 (lower). C, Cross-sections of mature seeds of DJY(left) and fse2 (right). E, F, Scanning electron microscopic (SEM) analysis of cross-sections of mature seeds of DJY (E) and fse2 (F). G-J, Semi-thin sections of DJY (G, H) and fse2 (I, J) endosperm at 10 days after pollination (DAP) stained with I2-KI. K, The 1000-grain weight of DJY and fse2. n=3. L, Grain length, width and thickness in DJY and fse2. n=20. All values are expressed as mean±SD. **means significant difference at 0.01 level (Student’s t-test).
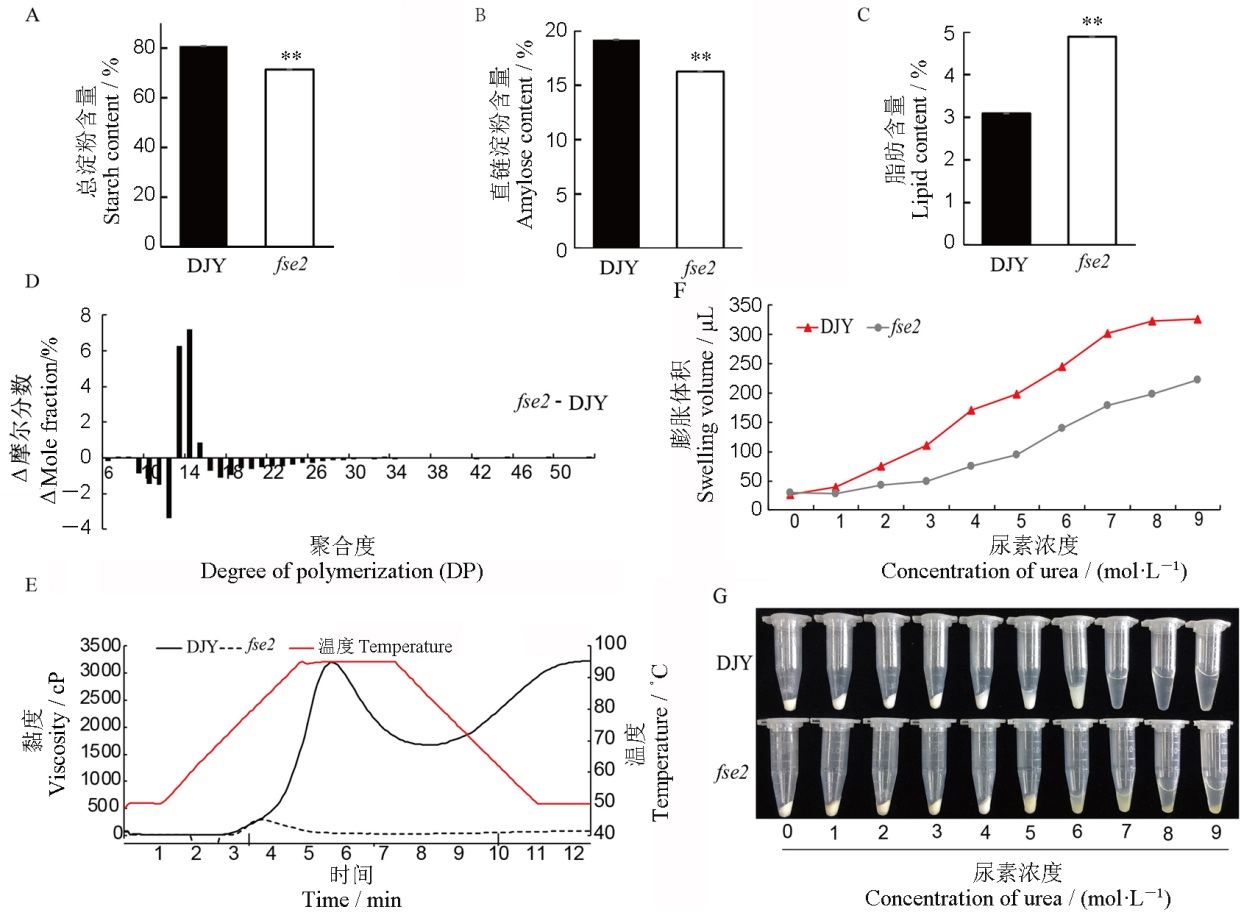
Fig. 2. Physicochemical characteristics of mature seeds of fse2 and its wild type. A~C, The contents of total starch (A), amylose (B), and lipid (C) in the endosperm of Dianjingyou(DJY) and fse2. n=3, Values are means±SD, Student’s t-test, **P<0.01. D, Amylopectin chain length distributions of DJY and fse2. E, Analysis of RVA characteristic of starch in DJY and fse2. F, The swollen volume of DJY and fse2 starch in urea solution (n=3). G, Starch expansion of DJY and fse2 in urea solutions.
| 试材 Test material | 最高黏度 Peak viscosity | 热浆黏度 Hot pasting viscosity | 崩解值Breakdown viscosity | 冷胶黏度 Cool pasting viscosity | 消减值 Setback viscosity | 峰值时间 Peak time / min | 糊化温度 Gelatinization temperature /℃ |
|---|---|---|---|---|---|---|---|
| DJY | 3197 | 1677 | 1520 | 3225 | 28 | 5.67 | 76.00 |
| fse2 | 293 | 34 | 259 | 83 | -210 | 3.80 | 75.95 |
Table 1 Analysis of RVA characteristic values of starch in wild type and fse2 mutant.
| 试材 Test material | 最高黏度 Peak viscosity | 热浆黏度 Hot pasting viscosity | 崩解值Breakdown viscosity | 冷胶黏度 Cool pasting viscosity | 消减值 Setback viscosity | 峰值时间 Peak time / min | 糊化温度 Gelatinization temperature /℃ |
|---|---|---|---|---|---|---|---|
| DJY | 3197 | 1677 | 1520 | 3225 | 28 | 5.67 | 76.00 |
| fse2 | 293 | 34 | 259 | 83 | -210 | 3.80 | 75.95 |
| 年份 Year | 透明种子数 Number of normal seeds | 粉质皱缩种子数 Number of floury and shrunken seeds | χ2(3:1) |
|---|---|---|---|
| 2017 | 681 | 208 | 1.508 |
| 2016 | 573 | 172 | 1.612 |
Table 2 Genetic analysis of fse2.
| 年份 Year | 透明种子数 Number of normal seeds | 粉质皱缩种子数 Number of floury and shrunken seeds | χ2(3:1) |
|---|---|---|---|
| 2017 | 681 | 208 | 1.508 |
| 2016 | 573 | 172 | 1.612 |
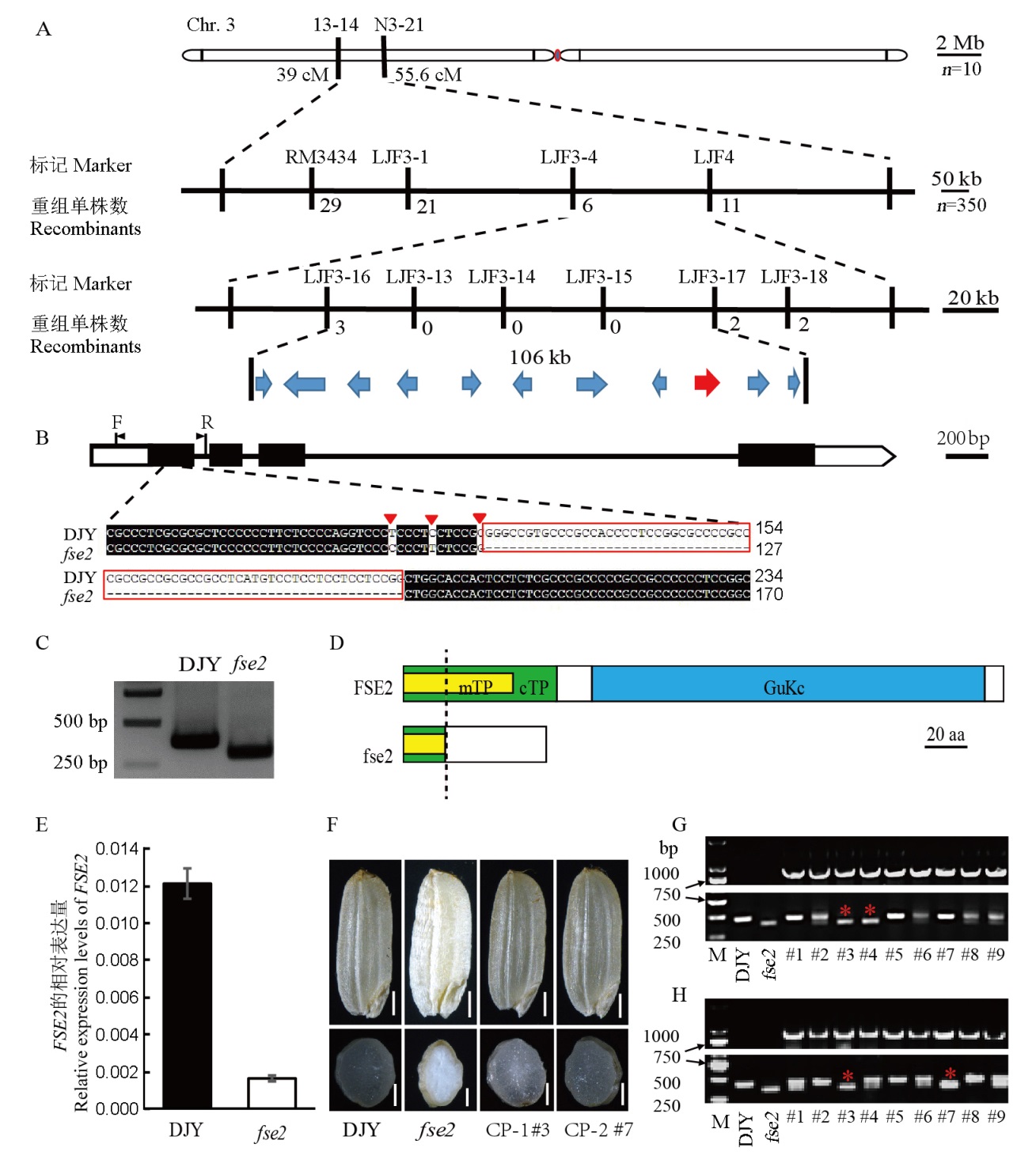
Fig. 3. Map-based cloning of FSE2. A, Map-based cloning of the FSE2 locus. The FSE2 locus was mapped to a 106 kb region by markers LJF3-16 and LJF3-17 on the short arm of chromosome 3, which contains 11 predicted open reading frames (ORFs). B, The structure of Os03g0320900 and the mutation site. Three nucleotide substitutions (red triangles) and a 70 bp deletion (red box) in the sequence are indicated. The genetic background was identified with the primer pair F/R in transgenic lines. C, PCR analysis of the 70 bp deletion in the genomic region of Os03g0320900. D, Target peptides and functional domain of FSE2 protein. A total of 68 amino acids in fse2, and only the first 20 amino acids in the N-terminus are consistent with the wild type (left of the dotted line). mTP, Mitochondrial transit peptide; cTP, Chloroplast transit peptide; aa, amino acids. E, The relative expression levels of developing seeds (10 days post-pollination) in DJY and fse2. F, Positive transgenic seeds with homozygous fse2 background (CP-1 #3 and CP-2 #7) showed transparent endosperm. CP-1 and CP-2 are two positive transgenic lines with heterozygous background. Bar=1 cm. G-H, PCR analyses of the transparent seeds from CP-1 and CP-2 lines. The upper panels in G and H represent positive transgenic individuals. The lower panels of G and H show the backgrounds of these positive transgenic individuals. The red asterisks indicate the positive transgenic seeds with homozygous fse2 background. DJY, Dianjingyou 1.
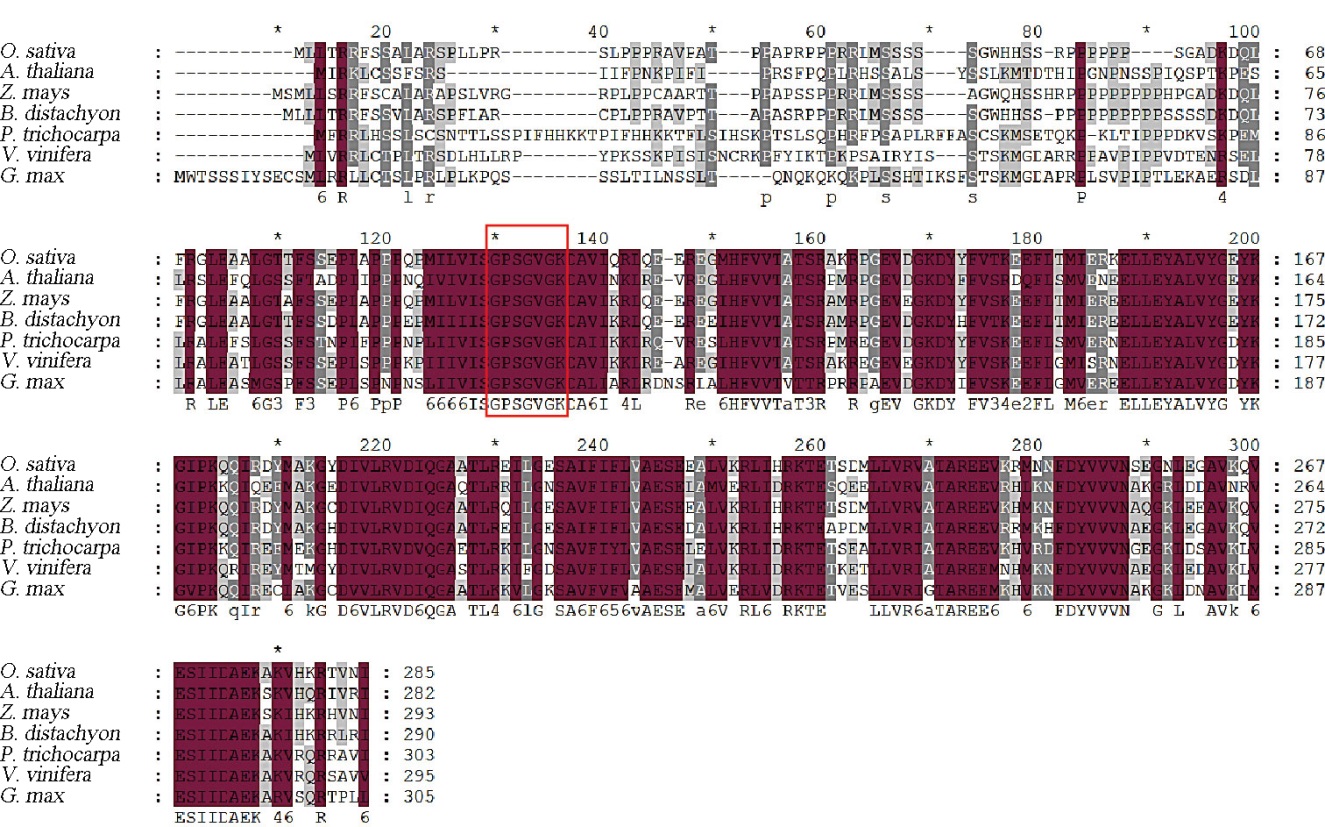
Fig. 4. Protein sequence alignment of FSE2 and its homologs. The red box indicates the catalytic site of GK; GenBank protein accession number are as follows O. sativa, XP_051628708.1; A. thaliana, NP_566276.1; Z. mays, NP_001149581.1; B. distachyon, XP_003561683.1; P. trichocarpa, XP_006380050.1; V. vinifera, XP_002279802.1; G. max, XP_003542897.2.
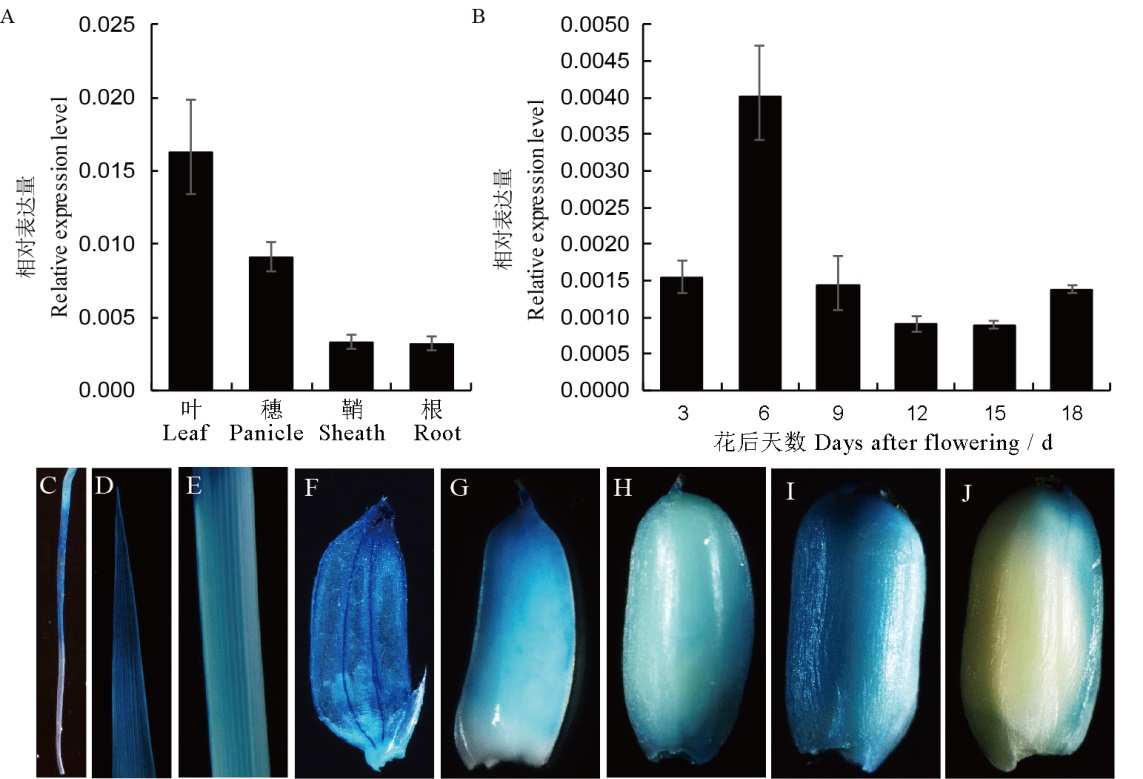
Fig. 5. The expression patterns of FSE2. A, Expression levels of FSE 2 in leaf, panicle, sheath and root of wild type. B, Expression levels of FSE2 in the developing endosperms of 3, 6, 9, 12, 15 and 18 days after flowering. C-F, GUS staining patterns in root (C), leaf (D), sheath (E) and panicle (F); G-J, GUS staining patterns in developing endosperm of 6 (G), 9 (H), 12 (I) and 15 days after flowering (J).
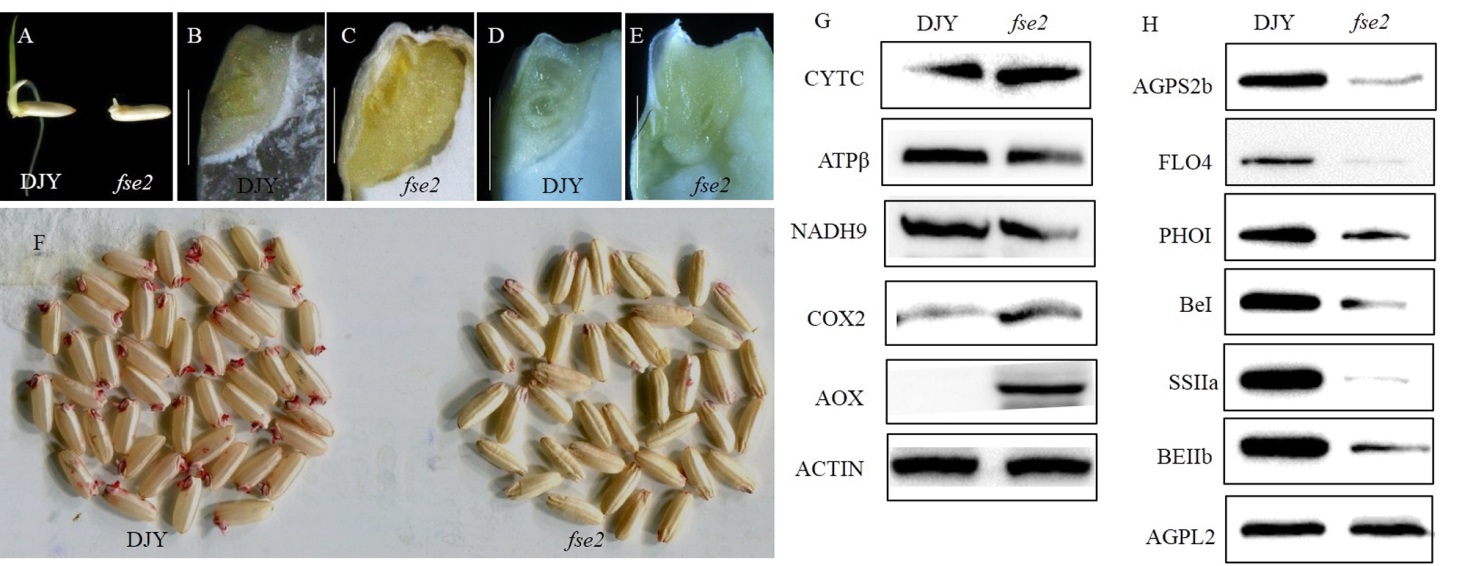
Fig. 6. The abnormal development of embryo and endosperm in the fse2 mutant. A, Comparison of the seed of wild type and fse2 after germinating for two days. B,C, Embryos of wild type and fse2 mutant seeds after imbibition (30℃, 9 h). Bars=1 mm. D, E, Developing embryos of 15 DAP seeds of wild type and fse2. Bars=1 mm. F, The determination of seed viability by TTC staining. G, Western blotting analysis of mitochondrial proteins in mature seeds of wild type and mutant. CytC, Cytochrome c biogenesis C; COX2, Cytochrome c oxidase subunit 2; NADH9, NADH dehydrogenase subunit 9; ATPβ, ATP synthase F0 subunit 6; AOX, Alternative oxidase. H, Western blotting analysis of starch synthesis enzymes in mature seeds of wild type and mutant. AGPL2, ADP-glucose pyrophosphorylase large subunit 2; AGPS2b, ADP-glucose pyrophosphorylase small subunit 2b; PPDKB, Cytosolic pyruvate orthophosphate dikinase B; PHOⅠ, Plastid phosphorylaseⅠ; BEⅠ, Starch-branching enzyme; BEⅡb, Starch-branching enzymeⅡb; SSⅡa, Starch synthaseⅡa. ACTIN antibody was used as a loading control in G and H. DJY, Dianjingyou 1.
| [1] | Khush G S.What it will take to feed 5.0 billion rice consumers in 2030.Plant Mol Biol, 2005, 59(1): 1-6. |
| [2] | Zhou Z, Robards K, Heliwell S, Blanchard C.Composition and functional properties of rice.Int J Food Sci Technol, 2002, 37(8): 849-868. |
| [3] | Demirkesen I, Sumnu G, Sahin S.Image analysis of gluten-free breads prepared with chestnut and rice flour and baked in different ovens.Food Bioprocess Technol, 2013, 6(7): 1749-1758. |
| [4] | Patindol J, Wang Y J.Fine structures and physicochemical properties of starches from chalky and translucent rice kernels.J Agric Food Chem, 2003, 51(9): 2777-2784. |
| [5] | Martin C, Smith A M.Starch biosynthesis.Plant Cell, 1995, 7(7): 971-985. |
| [6] | Nakamura Y.Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: Rice endosperm as a model tissue.Plant & Cell Physiol, 2002, 43(7): 718-725. |
| [7] | Hirose T, Terao T.A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta, 2004, 220(1): 9-16. |
| [8] | Ball S G, Morell M K.From bacterial glycogen to starch: Understanding the biogenesis of the plant starch granule.Annu Rev Plant Biol, 2003, 54(1): 207-233. |
| [9] | Colleoni C, Dauvillée D, Mouille G, Morell M, Samuel M, Slomiany M C, Lienard L, Wattebled F, d’Hulst C, Ball S. Biochemical characterization of the Chlamydomonas reinhardtii α-1,4 glucanotransferase supports a direct function in amylopectin biosynthesis. Plant Physiol, 1999, 120(4): 1005-1014. |
| [10] | Dauvillée D, Chochois V, Steup M, Haebel S, Eckermann N, Ritte G, Ral J P, Colleoni C, Hicks G, Wattebled F O, Deschamps P, d’Hulst C O, Liénard L, Cournac L O, Putaux J L O, Dupeyre D, Ball S G O. Plastidial phosphorylase is required for normal starch synthesis in Chlamydomonas reinhardtii. Plant J, 2006, 48(2): 274-285. |
| [11] | Dong X, Zhang D, Liu J, Liu Q Q, Liu H, Tian L, Jiang L, Qu le Q. Plastidial disproportionating enzyme participates in starch synthesis in rice endosperm by transferring maltooligosyl groups from amylose and amylopectin to amylopectin.Plant Physiol, 2015, 169(4): 2496-2512. |
| [12] | Schupp N, Ziegler P.The relation of starch phosphorylases to starch metabolism in wheat.Plant & Cell Physiol, 2004, 45(10): 1471-1484. |
| [13] | Satoh H, Shibahara K, Tokunaga T, Nishi A, Tasaki M, Hwang S K, Okita T W, Kaneko N, Fujita N, Yoshida M, Hosaka Y, Sato A, Utsumi Y, Ohdan T, Nakamura Y.Mutation of the plastidial a-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm.Plant Cell, 2008, 20(7): 1833-1849. |
| [14] | Pfeilmeier S, Saur I M, Rathjen J P, Zipfel C, Malone J G.High levels of cyclic-di-GMP in plant-associated Pseudomonas correlate with evasion of plant immunity.Mol Plant Pathol, 2016, 17(4): 521-531. |
| [15] | Zrenner R, Stitt M, Sonnewald U, Boldt R.Pyrimidine and purine biosynthesis and degradation in plants.Annu Rev Plant Biol, 2006, 57: 805-836. |
| [16] | Green R, Noller H F.Ribosomes and translation.Annu Rev Biochem, 1997, 66: 679-716. |
| [17] | Sumita K, Lo Y H, Takeuchi K, Senda M, Kofuj S, Ikeda Y, Terakawa J, Sasaki M, Yoshino H, Majd N, Zheng Y X, Kahoud E R, Yokota T, Emerling B M, Asara J M, Ishida T, Locasale J W, Daikoku T, Anastasiou D, Senda T, Sasaki A T.The lipid kinase PI5P4Kβ is an intracellular GTP sensor for metabolism and tumorigenesis.Mol Cell, 2016, 61(2): 187-198. |
| [18] | Caro L G, Palade G E.Protein synthesis, storage, and discharge in the pancreatic exocrine cell. An autoradiographic study. J Cell Biol, 1946, 20(3): 473-495. |
| [19] | Havel P J.Control of energy homeostasis and insulin action by adipocyte hormones: Leptin, acylation stimulating protein, and adiponectin.Curr Opin Lipidol, 2002, 13(1): 51-59. |
| [20] | Stasolla C, Katahira R, Thorpe T A, Ashihara H.Purine and pyrimidine nucleotide metabolism in higher plants.J Plant Physiol, 2003, 160(11): 1271-1295. |
| [21] | Gaidarov I O, Suslov O N, Abdulaev N G.Enzymes of the cyclic GMP metabolism in Bovine retina: I. Cloning and expression of the gene for guanylate kinase. FEBS Lett, 1993, 335(1): 81-84. |
| [22] | Brady W A, Kokoris M S, Fitzgibbon M, Black M E.Cloning, characterization, and modeling of mouse and human guanylate kinases.J Biol Chem, 1996, 271(28): 16734-16740. |
| [23] | Stolworthy T S, Krabbenhoft E, Black M E.A novel Escherichia coli strain allows functional analysis of guanylate kinase drug resistance and sensitivity.Anal Biochem, 2003, 322(1): 40-47. |
| [24] | Beck B J, Huelsmeyer M, Paul S, Downs D M.A mutation in the essential gene gmk(encoding guanylate kinase) generates a requirement for adenine at low temperature in Salmonella enteric. J Bacteriol, 2003, 185(22): 6732-6735. |
| [25] | Gentry D, Bengra C, Ikehara K, Cashel M.Guanylate kinase of Escherichia coli K-12. J Biol Chem, 1993, 268(19): 14316-14321. |
| [26] | Konrad M.Cloning and expression of the essential gene for guanylate kinase from yeast.J Biol Chem, 1992, 267(36): 25652-25655. |
| [27] | Ray B D, Jarori G K, Raghunathan V, Yan H, Rao B D N. Conformations of nucleotides bound to wild type and Y78 F mutant yeast guanylate kinase: Proton two-dimensional transferred NOESY measurements. Biochemistry, 2005, 44(42): 13762 13770. |
| [28] | Kumar V.Cloning and sequence analysis of lily and tobacco guanylate kinases.Mol Biol Rep, 2000, 27(1): 45 49. |
| [29] | Kumar V, Spangenberg O, Konrad M.Cloning of the guanylate kinase homologues AGK-1 and AGK-2 from Arabidopsis thaliana and characterization of AGK-1. Eur J Biochem, 2000, 267(2): 606-615 |
| [30] | Sugimoto H, Kusumi K, Tozawa Y, Yazaki H, Kishimoto N, Kikuchi S, Iba K.The virescent-2 mutation inhibits translation of plastid transcripts for the plastid genetic system at an early stage of chloroplast differentiation. Plant & Cell Physiol, 2004, 45(8): 985-996. |
| [31] | Sugimoto H, Kusumi K, Noguchi K, Yano M, Yoshimura A, Iba K.The rice nuclear gene,VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J, 2007, 52(3): 512-527. |
| [32] | Kang H G, Park S, Matsuoka M, An G.White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J, 2005, 42(6): 901-911. |
| [33] | Peng C, Wang Y, Liu F, Ren Y, Zhou K, Lv J, Zheng M, Zhao M, Zhao S, Zhang L, Wang C, Jiang L, Zhang X, Guo X, Wan J M.FLOURY ENDOSPERM 6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. Plant J, 2014, 77(6): 917-930. |
| [34] | Nishi A, Nakamura Y, Tanaka N, Satoh H.Biochemical and genetic analysis of the effects of Amylose-Extender mutation in rice endosperm. Plant Physiol, 2001, 127(2): 459-472 |
| [35] | Ohdan T, Francisco P B Jr, Sawada T, Hirose T, Saltoh H, Nakamura Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot, 2005, 56(422): 3229-3244. |
| [36] | Akihiro T, Mizuno K, Fujimura T.Gene expression of ADP-glucose pyrophosphorylase and starch contents in rice cultured cells are cooperatively regulated by sucrose and ABA.Plant & Cell Physiol, 2005, 46(6): 937-946. |
| [37] | Finnegan P M, Soole K L, Umbach A L.Alternative mitochondrial electron transport proteins in higher plants. //Day D A, Millar A H, Whelan J. Plant Mitochondria: From Genome to Function. The Netherlands: Springer, 2004: 163-230. |
| [38] | Selinshi J, Hartmann A, Kordes A, Deckers-Hebestreit G, Whelan J, Scheibe R.Analysis of post-translational activation of alternative oxidase isoforms.Plant Physiol, 2017, 174(4): 2113-2127. |
| [39] | Zhang Y F, Suzuki M, Sun F, Tan B C.The mitochondrion-targeted PENTATRICOPEPTIDE REPEAT78 protein is required fornad5 mature mRNA stability and seed development in maize. Mol Plant, 2017, 10(10): 1321-1333 |
| [40] | Li X J, Zhang Y F, Hou M M, Sun F, Shen Y, Xiu Z H, Wang X, Chen Z L, Sun S S, Small I, Tan B C.Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize(Zea mays) and rice, 2014, 79(5): 797-809. |
| [41] | Liu Y J, Xiu Z H, Meeley R, Tan B C.Empty Pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell, 2013, 25(3): 868-883. |
| [42] | Yang Y Z, Ding S, Wang H C, Sun F, Huang W L, Song S, Xu C, Tan B C.The pentatricopeptide repeat protein EMP9 is required for mitochondrial ccmB and rps4 transcript editing, mitochondrial complex biogenesis and seed development in maize. New Phytol, 2017, 214(2): 782-795. |
| [43] | Cai M J, Li S Z, Sun F, Sun Q, Zhao H, Ren X, Zhao Y, Tan B C, Zhang Z, Qiu F.Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. Plant J, 2017, 91(1): 132-144. |
| [44] | Ren X M, Pan Z Y, Zhao H L, Zhao J L, Cai M J, Li J, Zhang Z X, Qiu F Z.EMPTY PERICARP11 serves as a factor for splicing of mitochondrial nad1 intron and is required to ensure proper seed development in maize. J Exp Bot, 2017, 68(16): 4571-4581. |
| [45] | Jiang P F, Wang S L, Jiang H Y, Cheng B J, Wu K Q, Ding Y.The COMPASS-like complex promotes flowering and panicle branching in rice. Plant Physiol, 176(4): 01749.2017. DOI:10.1104/pp.17.01749. |
| [46] | Minkenberg B, Xie K, Yang Y N.Discovery of rice essential genes by characterizing a CRISPR-edited mutation of closely related rice MAP kinase genes.Plant J, 2017, 89(3): 636-648. |
| [47] | Huang X, Peng X, Sun M X.OsGCD1 is essential for rice fertility and required for embryo dorsal-ventral pattern formation and endosperm development.New Phytol, 2017, 215(9): 1039-1058. |
| [1] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [2] | ZHOU Tian, WU Shaohua, KANG Jianhong, WU Hongliang, YANG Shenglong, WANG Xingqiang, LI Yu, HUANG Yufeng. Effects of Planting Patterns on Starch Content and Activities of Key Starch Enzymes in Rice Grains [J]. Chinese Journal OF Rice Science, 2024, 38(3): 303-315. |
| [3] | WU Ziniu, HE Limei, XIONG Ying, CHEN Kairui, YANG Zhiyuan, SUN Yongjian, LÜ Xu, MA Jun. Effect of Nitrogen Fertilizer Topdressing for Panicle Differentiation on Grain Filling of Hybrid indica Rice and Its Relationship with the Activities of Key Enzymes for Starch Synthesis [J]. Chinese Journal OF Rice Science, 2024, 38(1): 48-56. |
| [4] | CHEN Liming, YANG Taotao, XIONG Ruoyu, TAN Xueming, HUANG Shang, ZENG Yongjun, PAN Xiaohua, SHI Qinghua, ZHANG Jun, ZENG Yanhua. Effect of Free-air Temperature Increasing on Activities of Enzymes Involved in Starch Synthesis and Accumulation of Double-cropping indica Rice [J]. Chinese Journal OF Rice Science, 2023, 37(2): 166-177. |
| [5] | CHEN Hongyang, JIA Yan, ZHAO Hongwei, QU Zhaojun, WANG Xinpeng, DUAN Yuyang, YANG Rui, BAI Xu, WANG Changcheng. Effects of Low Temperature Stress During Grain Filling on Starch Formation and Accumulation of Superior and Inferior Grains in Rice [J]. Chinese Journal OF Rice Science, 2022, 36(5): 487-504. |
| [6] | LIANG Cheng, XIANG Xunchao, ZHANG Ouling, YOU Hui, XU Liang, CHEN Yongjun. Analyses on Agronomic Traits and Genetic Characteristics of Two New Plant-architecture Lines in Rice [J]. Chinese Journal OF Rice Science, 2022, 36(2): 171-180. |
| [7] | Yujun ZHU, Ziwei ZUO, Zhenhua ZHANG, Yeyang FAN. A New Approach for Fine-mapping and Map-based Cloning of Minor-Effect QTL in Rice [J]. Chinese Journal OF Rice Science, 2021, 35(4): 407-414. |
| [8] | Yali ZHENG, Linchuang YU, Xiaoxiao AN, Xinle CHENG, Lijun REN, Zilong SU, Xiaoya ZHENG, Tao LAN. Identification of a Knockout Mutant of OsWOX3B Gene in Rice (Oryza sativa L.) [J]. Chinese Journal OF Rice Science, 2021, 35(2): 112-120. |
| [9] | Yiwei KANG, Yuyu CHEN, Yingxin ZHANG. Research Progress and Breeding Prospects of Grain Size Associated Genes in Rice [J]. Chinese Journal OF Rice Science, 2020, 34(6): 479-490. |
| [10] | DU Yimo, PAN Tian, TIAN Yunlu, LIU Shijia, LIU Xi, JIANG Ling, ZHANG Wenwei, WANG Yihua*,WAN Jianmin . Phenotypic Analysis and Gene Cloning of Rice Floury Endosperm Mutant fse4 [J]. Chinese Journal of Rice Science, 2019, 33(6): 499-512. |
| [11] | Yanhua CHEN, Yaliang WANG, Defeng ZHU, Qinghua SHI, Huizhe CHEN, Jing XIANG, Yikai ZHANG, Yuping ZHANG. Mechanism of Exogenous Brassinolide in Alleviating High Temperature Injury at Panicle Initiation Stage in Rice [J]. Chinese Journal OF Rice Science, 2019, 33(5): 457-466. |
| [12] | Chen CHENG, Yongjun ZENG, Huihuang CHENG, Xueming TAN, Qingyin SHANG, Yanhua ZENG, Qinghua SHI. Effects of Different Temperature from Full Heading to Milking on Grain Filling Stage on Grain Hormones concentrations, Activities of Enzymes Involved in Starch Synthesis and Accumulation in Rice Nanjing 9108 [J]. Chinese Journal OF Rice Science, 2019, 33(1): 57-67. |
| [13] | Tao SUN, Laga TONG, Shuyu ZHAO, Haiwei WANG, Yunfei HAN, Zhongchen ZHANG, Zhengxun JIN. Effects of Nitrogen Fertilizer Application on Starch Quality, Activities and Gene Expression Levels of Related Enzymes in Rice Endosperm [J]. Chinese Journal OF Rice Science, 2018, 32(5): 475-484. |
| [14] | Xichun ZHANG, Feifei LU, Yusong LÜ, Rongjian LUO, Guiai JIAO, Yawen WU, Shaoqing TANG, Peisong HU, Xiangjin WEI. Identification and Gene Mapping-based Clone of Two Chalkiness Mutants in Rice [J]. Chinese Journal OF Rice Science, 2017, 31(6): 568-579. |
| [15] | Yaling CHEN, Jinsong BAO. Progress in Structures, Functions and Interactions of Starch Synthesis Related Enzymes in Rice Endosperm [J]. Chinese Journal OF Rice Science, 2017, 31(1): 1-12. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||