
Chinese Journal OF Rice Science ›› 2015, Vol. 29 ›› Issue (2): 126-134.DOI: 10.3969/j.issn.1001-7216.2015.02.003
• Orginal Article • Previous Articles Next Articles
Xiao-jing LI1, Duo-duo XU1, Yi-min XU1, Kai-en ZHAI1, Yao-long YANG2, Jian-wei PAN1, Yu-chun RAO1,2,*( )
)
Received:2014-07-27
Revised:2014-12-02
Online:2015-03-10
Published:2015-03-10
Contact:
Yu-chun RAO
李晓静1, 徐多多1, 徐益敏1, 翟开恩1, 杨窑龙2, 潘建伟1, 饶玉春1,2,*( )
)
通讯作者:
饶玉春
基金资助:CLC Number:
Xiao-jing LI, Duo-duo XU, Yi-min XU, Kai-en ZHAI, Yao-long YANG, Jian-wei PAN, Yu-chun RAO. Expression of OsBC88, a Rice Cellulose Synthase Catalytic Subunit Gene[J]. Chinese Journal OF Rice Science, 2015, 29(2): 126-134.
李晓静, 徐多多, 徐益敏, 翟开恩, 杨窑龙, 潘建伟, 饶玉春. 水稻纤维素合酶催化亚基的编码基因BC88的表达分析[J]. 中国水稻科学, 2015, 29(2): 126-134.
Add to citation manager EndNote|Ris|BibTeX
URL: http://www.ricesci.cn/EN/10.3969/j.issn.1001-7216.2015.02.003
| 引物名称 Primer | 引物序列(5'-3') Primer sequence(5'-3') |
|---|---|
| pBC88-2035-HindⅢ-F | CCGAAGCTTCTTTTCCATTTTCATATTTCCCTGCCATCA |
| pBC88-3896-BamHⅠ-R | GCCGGATCCGTGTGGGAGACGGTGGCGGGTTGGTTG |
| BC88-no ATG-XbaⅠ-F | GGCTCTAGAGAGGCGAGCGCCGGGCTGGTGGCCGG |
| BC88-stop codon-XbaⅠ-R | AGCTCTAGATCAGCAGTTGATGCCGCACTGCCTGACGTC |
| BC88-600-R | CTCAGACACCGGGTAGGGGTGGATGC |
| OsActin-Real-F | TGGCATCTCTCAGCACATTCC |
| OsActin-Real-R | TGCACAATGGATGGGTCAGA |
| OsBC-104-F | AGGTGTGCGAGATATGCGGCGACGAG |
| OsBC-213-R | CTCGTACTCGTAGCAGGGGCGGCACAC |
Table 1 Primer sequences for OsBC88 cloning and Real-time PCR.
| 引物名称 Primer | 引物序列(5'-3') Primer sequence(5'-3') |
|---|---|
| pBC88-2035-HindⅢ-F | CCGAAGCTTCTTTTCCATTTTCATATTTCCCTGCCATCA |
| pBC88-3896-BamHⅠ-R | GCCGGATCCGTGTGGGAGACGGTGGCGGGTTGGTTG |
| BC88-no ATG-XbaⅠ-F | GGCTCTAGAGAGGCGAGCGCCGGGCTGGTGGCCGG |
| BC88-stop codon-XbaⅠ-R | AGCTCTAGATCAGCAGTTGATGCCGCACTGCCTGACGTC |
| BC88-600-R | CTCAGACACCGGGTAGGGGTGGATGC |
| OsActin-Real-F | TGGCATCTCTCAGCACATTCC |
| OsActin-Real-R | TGCACAATGGATGGGTCAGA |
| OsBC-104-F | AGGTGTGCGAGATATGCGGCGACGAG |
| OsBC-213-R | CTCGTACTCGTAGCAGGGGCGGCACAC |
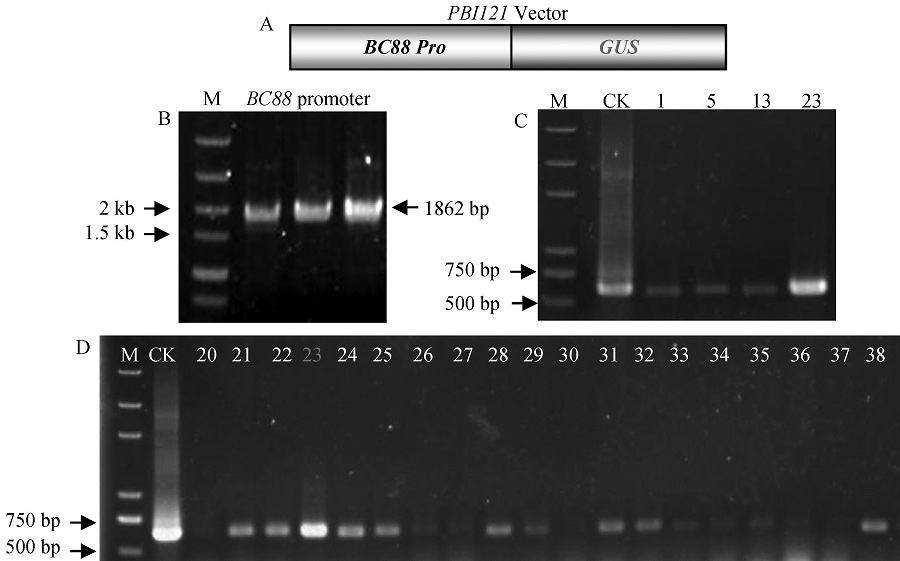
Fig.1. Construction of PBI121 BC88 pro :: GUS vector. A, Structure of PBI121 BC88 pro :: GUS vector; B, PCR products of BC88 promoter; C, PCR identification of positive bacterial suspension; D, PCR identification of positive bacterial colony; M, DNA marker; CK, Positive control.
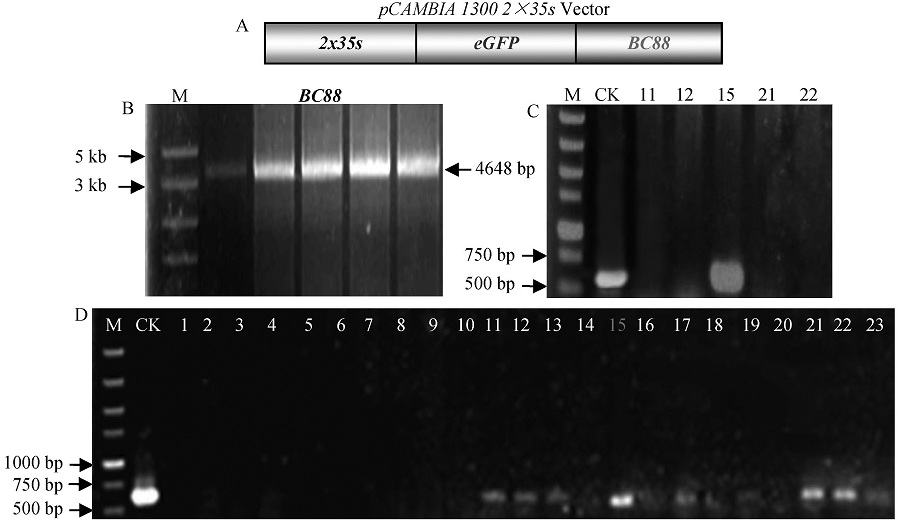
Fig. 2. Construction of pcambia 1300-2×35s:: eGFP: BC88 vector. A, Structure of pcambia 1300-2×35s:: eGFP: BC88 vector; B, PCR products of BC88; C, PCR identification of positive bacterial suspension; D, PCR identification of positive bacterial colony, M, DNA marker; CK, Positive control.
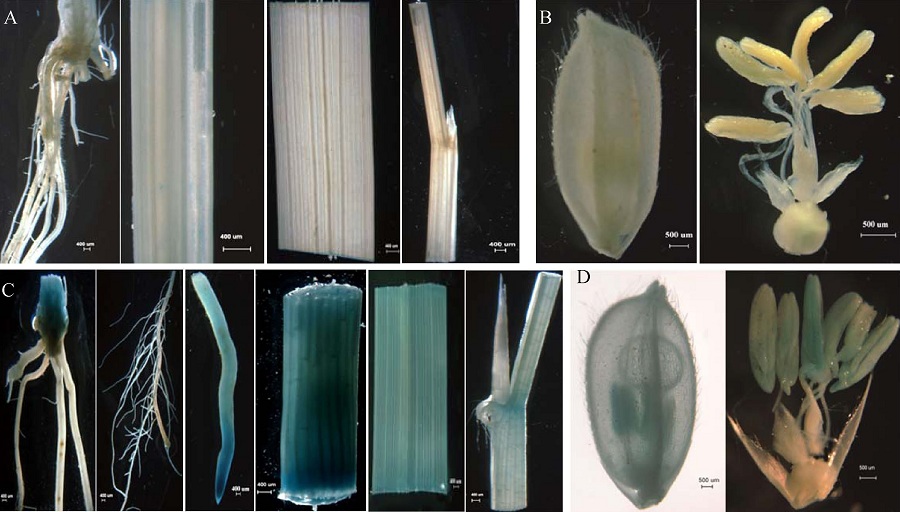
Fig. 3. Tissue expression analysis of OsBC88. A, GUS staining of root, culm, leaf, sheath of Nipponbare at seedling stage; B, GUS staining of flower of Nipponbare at flowering stage, serving as negative control; C, GUS staining of root, culm, sheath, leaf of pBI121: BC88 pro :: GUS transgenic lines at seedling stage; D, GUS staining of flower of pBI121:BC88 pro:: GUS transgenic lines at flowering stage.
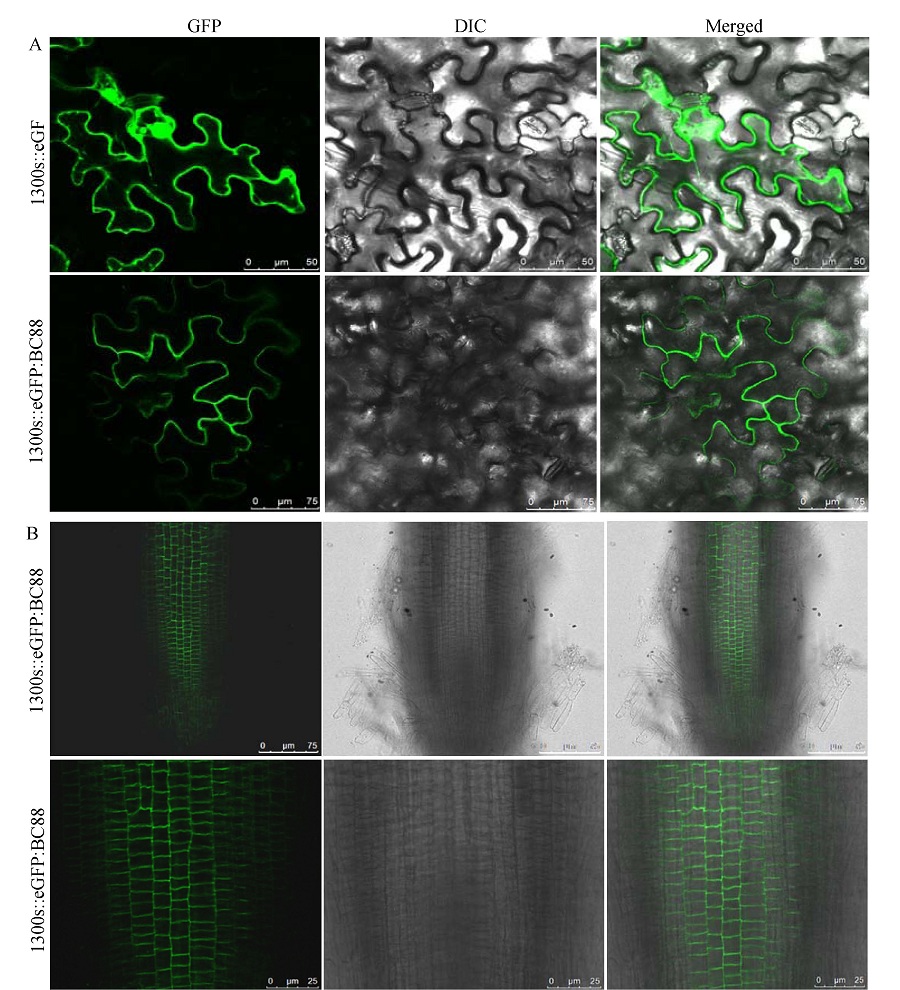
Fig. 5. Subcellular localization of BC88. A, Localization of 35S:: eGFP and 35S:: eGFP: BC88 in tobacco epidermal cells; B, Localization of 35S:: eGFP in rice root tip cells, the bottom is the amplification of the upside. GFP indicates the green fluorescence of proteins, DIC indicates the differential interference contrast phase, and Merged indicates the merging of GFP and DIC.
| [1] | Cosgrove D J.Relaxation in a high-stress environment: The molecular bases of extensible cell walls and cell enlargement.Plant Cell, 1997, 9(7): 1031-1041. |
| [2] | Kotilainen M, Helariutta Y, Mehto M, et al.GEG participates in the regulation of cell and organ shape during corolla and carpel development in Gerbera hybrida.Plant Cell, 1999, 11(6): 1093-1104. |
| [3] | Martin C, Bhatt K, Baumann K.Shaping in plant cells.Curr Opin Plant Biol, 2001, 4(6): 540-549. |
| [4] | Campbell M M, Sederoff R R.Variation in lignin content and composition: Mechanism of control and implications for the genetic improvement of plants.Plant Physiol, 1996, 110(1): 3-13. |
| [5] | Keegstra K. Plant Cell Walls.Plant Physiol, 2010, 154(2): 483-486. |
| [6] | Cosgrove D J.Growth of the plant cell wall.Nat Rev Mol Cell Biol, 2005, 6(11): 850-861. |
| [7] | Crowell E F, Gonneau M, Vernhettes S, et al.Regulation of anisotropic cell expansion in higher plants.C R Biol, 2010, 333(4): 320-324. |
| [8] | 汪海峰, 朱军莉, 刘建新, 等.饲喂脆茎全株水稻对生长肥育猪生长性能、养分消化和胴体品质的影响.畜牧兽医学报, 2005, 36(11): 1139-1144. |
| [9] | Carpita N, McCann M. The cell wall//Buchanan R B, Gruissem W, Jones R L. Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, 2000: 52-108. |
| [10] | Somerville C.Cellulose synthesis in higher plants.Annu Rev Cell Dev Biol, 2006, 22:53-78. |
| [11] | Taylor N G, Scheible W R, Cutler S, et al.The irregular xylern3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis.Plant Cell, 1999, 11(5): 769-779. |
| [12] | Lerouxel O, Cavalier D M, Liepman A H, et al.Biosynthesis of plant cell wall polysaccharides — a complex process.Curr Opin Plant Biol, 2006, 9(6): 621-630. |
| [13] | Baskin T I.Anisotropic expansion of the plant cell wall.Annu Rev Cell Dev Biol, 2005, 21: 203-222. |
| [14] | Pear J R, Kawagoe Y, Schreckengost W E, et al.Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase.Proc Natl Acad Sci USA, 1996, 93(22): 12637-12642. |
| [15] | Arioli T, Peng L, Betzner A S, et al.Molecular analysis of cellulose biosynthesis in Arabidopsis.Science, 1998, 279(5351): 717-720. |
| [16] | Fagard M, Desnoes T, Desprez T, et al.PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis.Plant Cell, 2001, 12(12): 2409-2423. |
| [17] | Taylor NG, Laurie S, Turner S R.Mutiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis.Plant Cell, 2000, 12(12): 2529-2539. |
| [18] | Tanaka K, Murata K, Yamazaki M, et al.Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall.Plant Physiol, 2003, 133(1): 73-83. |
| [19] | 童川, 童杰鹏, 孙出,等.水稻脆性基因的功能研究进展.分子植物育种, 2013, 11(2): 286-292. |
| [20] | Song X Q, Liu L F, Jiang Y J, et al.Disruption of secondary wall cellulose biosynthesis alters cadmium translocation and tolerance in rice plants.Mol Plant, 2013, 6(3): 768-780. |
| [21] | Rao Y C, Yang Y L, Xin D D, et al.Characterization and cloning of a brittle culm mutant ( bc88) in rice (Oryza sativa L.).Chin Sci Bull, 2013, 58(24): 3000-3006. |
| [22] | Kotake T, Aohara T, Hirano K, et al.Rice Brittle culm 6 encodes a dominant-negative form of CesA protein that perturbs cellulose synthesis in secondary cell walls.J Exp Bot, 2011, 62(6): 2053-2062. |
| [23] | 王超. 拟南芥网格蛋白轻链的功能分析.金华:浙江师范大学, 2012. |
| [24] | 沈金秋. 青藏高原野生大麦HsCIPKs基因克隆及其在水稻中的功能验证.金华:浙江师范大学, 2014. |
| [25] | Livak K J, Schmittgen T D.Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method.Methods, 2001, 25(4): 402-408. |
| [26] | Li Y H, Qian Q, Zhou Y H, et al.BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants.Plant Cell, 2003, 15(9): 2020-2031. |
| [27] | Dai X X, You C J, Chen G X, et al.OsBC1L4 encodes a COBRA-like protein that affects cellulose synthesis in rice.Plant Mol Biol, 2011, 75(4-5): 333-345. |
| [28] | Hirano K, Kotake T, Kamihara K, et al.Rice BRITTLE CULM 3 (BC3) encodes a classical dynamin OsDRP2B essential for proper secondary cell wall synthesis.Planta, 2010, 232(1): 95-108. |
| [29] | Aohara T, Kotake T, Kaneko Y, et al.Rice BRITTLE CULM 5 (BRITTLE NODE) is involved in secondary cell wall formation in the sclerenchyma tissue of nodes.Plant Cell Physiol, 2009, 50(11): 1886-1897. |
| [30] | Zhou Y H, Li S B, Qian Q, et al.BC10, a DUF266-containing and Golgi-located typeⅡ membrane protein, is required for cell-wall biosynthesis in rice (Oryza sativa L.).Plant J, 2009, 57(3): 446-462. |
| [31] | Zhang M, Zhang B C, Qian Q, et al. Brittle Culm 12, a dual-targeting kinesin-4 protein, controls cell-cycle progression and wall properties in rice.Plant J, 2010, 63(2): 312-328. |
| [32] | Zhang B C, Liu X L, Qian Q, et al.Golgi nucleotide sugar transporter modulates cell wall biosynthesis and plant growth in rice.PNAS, 2011, 108(12): 5110-5115. |
| [33] | Wu B, Zhang B C, Dai Y, et al.Brittle Culm 15 encodes a membrane-associated chitinase-like protein required for cellulose biosynthesis in rice.Plant Physiol, 2012, 159(4): 1440-1452. |
| [34] | Yan C, Yan S, Zeng X H, et al.Fine mapping and isolation of Bc7(t), allelic to OsCesA4.J Genet Genom, 2007, 34(11): 1019-1027. |
| [35] | Zhang B C, Deng L W, Qian Q, et al.A missense mutation in the transmembrane domain of CESA4 affects protein abundance in the plasma membrane and results in abnormal cell wall biosynthesis in rice.Plant Mol Biol, 2009, 71(4-5): 509-524. |
| [36] | Kimura S, Laosinchai W, Itoh T, et al.Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis.Plant Cell, 1999, 11(11): 2075-2086. |
| [37] | Peng L, Kawagoe Y, Hogan P, et al.Sitosterol-β-glucoside as primer for cellulose synthesis in plants.Science, 2002, 295(5552): 147-150. |
| [38] | Taylor N G, Howells R M, Huttly A K, et al.Interactions among three distinct CesA proteins essential for cellulose synthesis.P Natl Acad Sci USA, 2003, 100(3): 1450-1455. |
| [39] | Wang D F, Yuan S J, Yin L, et al.A missense mutation in the transmembrane domain of CESA9 affects cell wall biosynthesis and plant growth in rice.Plant Sci, 2012, 196: 117-124. |
| [40] | Ellis C, Karafyllidis I, Wasternack C, et al.The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses.Plant Cell, 2002, 14(17): 1557-1566. |
| [41] | Xiong G Y, Li R, Qian Q, et al.The rice dynamin-related protein DRP2B mediates membrane trafficking and thereby plays a critical role in secondary cell wall cellulose biosynthesis.Plant J, 2010, 64(1): 56-70. |
| [1] | GUO Zhan, ZHANG Yunbo. Research Progress in Physiological,Biochemical Responses of Rice to Drought Stress and Its Molecular Regulation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 335-349. |
| [2] | WEI Huanhe, MA Weiyi, ZUO Boyuan, WANG Lulu, ZHU Wang, GENG Xiaoyu, ZHANG Xiang, MENG Tianyao, CHEN Yinglong, GAO Pinglei, XU Ke, HUO Zhongyang, DAI Qigen. Research Progress in the Effect of Salinity, Drought, and Their Combined Stresses on Rice Yield and Quality Formation [J]. Chinese Journal OF Rice Science, 2024, 38(4): 350-363. |
| [3] | XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance [J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374. |
| [4] | CHEN Mingliang, ZENG Xihua, SHEN Yumin, LUO Shiyou, HU Lanxiang, XIONG Wentao, XIONG Huanjin, WU Xiaoyan, XIAO Yeqing. Typing of Inter-subspecific Fertility Loci and Fertility Locus Pattern of indica-japonica Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 386-396. |
| [5] | DING Zhengquan, PAN Yueyun, SHI Yang, HUANG Haixiang. Comprehensive Evaluation and Comparative Analysis of Jiahe Series Long-Grain japonica Rice with High Eating Quality Based on Gene Chip Technology [J]. Chinese Journal OF Rice Science, 2024, 38(4): 397-408. |
| [6] | HOU Xiaoqin, WANG Ying, YU Bei, FU Weimeng, FENG Baohua, SHEN Yichao, XIE Hangjun, WANG Huanran, XU Yongqiang, WU Zhihai, WANG Jianjun, TAO Longxing, FU Guanfu. Mechanisms Behind the Role of Potassium Fulvic Acid in Enhancing Salt Tolerance in Rice Seedlings [J]. Chinese Journal OF Rice Science, 2024, 38(4): 409-421. |
| [7] | LÜ Zhou, YI Binghuai, CHEN Pingping, ZHOU Wenxin, TANG Wenbang, YI Zhenxie. Effects of Nitrogen Application Rate and Transplanting Density on Yield Formation of Small Seed Hybrid Rice [J]. Chinese Journal OF Rice Science, 2024, 38(4): 422-436. |
| [8] | HU Jijie, HU Zhihua, ZHANG Junhua, CAO Xiaochuang, JIN Qianyu, ZHANG Zhiyuan, ZHU Lianfeng. Effects of Rhizosphere Saturated Dissolved Oxygen on Photosynthetic and Growth Characteristics of Rice at Tillering Stage [J]. Chinese Journal OF Rice Science, 2024, 38(4): 437-446. |
| [9] | WU Yue, LIANG Chengwei, ZHAO Chenfei, SUN Jian, MA Dianrong. Occurrence of Weedy Rice Disaster and Ecotype Evolution in Direct-Seeded Rice Fields [J]. Chinese Journal OF Rice Science, 2024, 38(4): 447-455. |
| [10] | LIU Fuxiang, ZHEN Haoyang, PENG Huan, ZHENG Liuchun, PENG Deliang, WEN Yanhua. Investigation and Species Identification of Cyst Nematode Disease on Rice in Guangdong Province [J]. Chinese Journal OF Rice Science, 2024, 38(4): 456-461. |
| [11] | HU Li, YANG Fanmin, CHEN Weilan, YUAN Hua. Research Progress in Biological Functions of SPL Family Transcription Factors in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 223-232. |
| [12] | CHEN Haotian, QIN Yuan, ZHONG Xiaohan, LIN Chenyu, QIN Jinghang, YANG Jianchang, ZHANG Weiyang. Research Progress on the Relationship Between Rice Root, Soil Properties and Methane Emissions in Paddy Fields [J]. Chinese Journal OF Rice Science, 2024, 38(3): 233-245. |
| [13] | MIAO Jun, RAN Jinhui, XU Mengbin, BO Liubing, WANG Ping, LIANG Guohua, ZHOU Yong. Overexpression of RGG2, a Heterotrimeric G Protein γ Subunit-Encoding Gene, Improves Drought Tolerance in Rice [J]. Chinese Journal OF Rice Science, 2024, 38(3): 246-255. |
| [14] | YIN Xiaoxiao, ZHANG Zhihan, YAN Xiulian, LIAO Rong, YANG Sijia, Beenish HASSAN, GUO Daiming, FAN Jing, ZHAO Zhixue, WANG Wenming. Signal Peptide Validation and Expression Analysis of Multiple Effectors from Ustilaginoidea virens [J]. Chinese Journal OF Rice Science, 2024, 38(3): 256-265. |
| [15] | ZHU Yujing, GUI Jinxin, GONG Chengyun, LUO Xinyang, SHI Jubin, ZHANG Haiqing, HE Jiwai. QTL Mapping for Tiller Angle in Rice by Genome-wide Association Analysis [J]. Chinese Journal OF Rice Science, 2024, 38(3): 266-276. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||