
中国水稻科学 ›› 2024, Vol. 38 ›› Issue (4): 364-374.DOI: 10.16819/j.1001-7216.2024.231215
许丹洁1,2,3, 林巧霞1,2,3, 李正康1,2,3, 庄小倩1,2,3, 凌宇1,2,3, 赖美玲1,2,3, 陈晓婷1,2,3,*( ), 鲁国东2
), 鲁国东2
收稿日期:2023-12-15
修回日期:2024-03-10
出版日期:2024-07-10
发布日期:2024-07-11
通讯作者:
*email: xiaotingchen@fafu.edu.cn
基金资助:
XU Danjie1,2,3, LIN Qiaoxia1,2,3, LI Zhengkang1,2,3, ZHUANG Xiaoqian1,2,3, LING Yu1,2,3, LAI Meiling1,2,3, CHEN Xiaoting1,2,3,*( ), LU Guodong2
), LU Guodong2
Received:2023-12-15
Revised:2024-03-10
Online:2024-07-10
Published:2024-07-11
Contact:
*email: xiaotingchen@fafu.edu.cn
摘要:
【目的】 茉莉酸(Jasmonic acid, JA)在植物生长发育和应对(非)生物胁迫中发挥重要功能,12-氧-植物二烯酸还原酶(12-oxo-phytodienoic acid reductase, OPR)是JA生物合成途径中的关键酶。创制OsOPR10转基因水稻,进行稻瘟病和白叶枯病抗性分析,揭示OsOPR10调控水稻抗病的分子机制,有望为水稻抗病育种提供种质资源。【方法】 构建OsOPR10的CRISPR/Cas9敲除载体和过表达载体,并分别利用农杆菌介导转化野生型水稻日本晴(Nipponbare, NPB),获得OsOPR10敲除和过表达转基因水稻。对转基因后代进行抗病性、活性氧(ROS)爆发、水杨酸(salicylic acid, SA)与茉莉酸(JA)途径基因表达模式分析。利用激光共聚焦显微镜观察OsOPR10的亚细胞定位,酵母双杂交实验和荧光素酶互补实验获取OsOPR10的互作蛋白。【结果】 获得OsOPR10纯合的敲除和过表达植株。接菌试验结果表明,OsOPR10过表达植株对稻瘟病和白叶枯病抗性更强。在几丁质(chitin)和细菌鞭毛蛋白(flg22)诱导后,OsOPR10过表达植株的ROS积累量明显高于野生型。qRT-PCR结果显示,在接种稻瘟菌12 h后,OsOPR10过表达植株中JA通路基因(OsAOS2、OsAOC)和SA通路基因(OsPR1a、OsPAL1)较NPB表达上调。亚细胞定位结果显示,OsOPR10蛋白定位于叶绿体。通过酵母双杂交试验获取OsOPR10的互作蛋白OsCYP28。【结论】 OsOPR10受稻瘟病菌和白叶枯菌以及外源茉莉酸甲酯与SA诱导。OsOPR10参与了病原菌分子模式触发的免疫途径(PTI),并且正调控水稻对稻瘟病和白叶枯病的抗性。此外,OsOPR10可能通过JA和SA途径介导水稻抗病反应。OsOPR10蛋白定位于叶绿体,与OsCYP28蛋白存在互作。
许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374.
XU Danjie, LIN Qiaoxia, LI Zhengkang, ZHUANG Xiaoqian, LING Yu, LAI Meiling, CHEN Xiaoting, LU Guodong. OsOPR10 Positively Regulates Rice Blast and Bacterial Blight Resistance[J]. Chinese Journal OF Rice Science, 2024, 38(4): 364-374.
| 引物名称 Primer name | 序列(5’→3’) Primer sequence(5’→3’) | 用途 Purpose |
|---|---|---|
| OsOPR10-F | ATGAAGTCAAGTCCAAATCAGCTG | CDS扩增 |
| OsOPR10-R | ACGAGTTGCTAGTTCTGAATTCTGAT | CDS amplification |
| OsOPR10-OE-F | CTTCTGCAGCCCGGGGGATCCATGAAGTCAAGTCCAAATCAGCTG | 过表达载体构建 |
| OsOPR10-OE-R | CGATCGGGGAAATTCGGATCCTCAAAGATCTTCTTCAGAAATCAACTTTTGTTCACGAGTTGCTAGTTCTGAATTCTGAT | Construction of overexpression vector |
| UbiP-seq | TTTTAGCCCTGCCTTCATACGC | 过表达载体测序 |
| NosR-seq | AGACCGGCAACAGGATTCAATC | Overexpression vector sequencing |
| OsOPR10-gRT1 | CGGCCGCCTGCTTCCGTTCGTTTTAGAGCTAGAAAT | CRISPR/Cas9第一个靶点扩增 |
| OsOPR10-OsU3T1 | GAACGGAAGCAGGCGGCCGTGCCACGGATCATCTGC | Amplification of the first CRISPR/Cas9 target |
| OsOPR10-gRT2 | AGATCGGCGCGCACCGCCTGTTTTAGAGCTAGAAAT | CRISPR/Cas9第二个靶点扩增 |
| OsOPR10-U6aT2 | AGGCGGTGCGCGCCGATCTCGGCAGCCAAGCCAGCA | Amplification of the second CRISPR/Cas9 target |
| OsOPR10-T1-F | AGAGAACCGGTGCCGCTTC | CRISPR/Cas9第一个靶点验证 |
| OsOPR10-T1-R | GGTCGGAGTCGTGGCAGTC | First CRISPR/Cas9 target validation |
| Actin-QF | GAGTATGATGAGTCGGGTCCAG | Actin 定量 PCR |
| Actin-QR | ACACCAACAATCCCAAACAGAG | qRT-PCR of Actin |
| OsUG-F | TTCTGGTCCTTCCACTTTCAG | 泛素融合蛋白定量PCR |
| OsUG-R | ACGATTGATTTAACCAGTCCATGA | qRT-PCR of ubiquitin fusion protein OsUG |
| MoPot2-F | ACGACCCGTCTTTACTTATTTGG | MoPot2 定量 PCR |
| MoPot2-R | AAGTAGCGTTGGTTTTGTTGGAT | qRT-PCR of MoPot2 |
| OsOPR10-BD-F | TCAGAGGAGGACCTGCATATGATGAAGTCAAGTCCAAATCAGCTG | OsOPR10-BD载体构建 |
| OsOPR10-BD-R | TCGACGGATCCCCGGGAATTCTCAACGAGTTGCTAGTTCTGAATTC | Construction of OsOPR10-BD vector |
| OsCYP28-AD-F | GTACCAGATTACGCTCATATGATGGTGTTGCCTTCATCAAA | OsCYP28-AD载体构建 |
| OsCYP28-AD-R | ATGCCCACCCGGGTGGAATTCTTATGGCAGACTTGGAGGGG | Construction of OsCYP28-AD vector |
| OsOPR10-pGDG-F | gcttcgaattctgcagtcgacATGAAGTCAAGTCCAAATCAGCTG | OsOPR10-pGDG载体构建 |
| OsOPR10-pGDG-R | ttatctagatccggtggatccTCAACGAGTTGCTAGTTCTGAATTC | Construction of OsOPR10-pGDG vector |
| OsOPR10-PHF223-F | CGGGATCGATGAAGTCAAGTCCAAATCAG | OsOPR10-PHF223载体构建 |
| OsOPR10-PHF223-R | GGGGTACCACGAGTTGCTAGTTCTGAATT | Construction of OsOPR10-PHF223 vector |
| OsPR1a-QF | CGTCTTCATCACCTGCAACTACTC | OsPR1a 定量 PCR |
| OsPR1a-QR | CATGCATAAACACGTAGCATAGCA | qRT-PCR of OsPR1a |
| OsAOS2-QF | CAATACGTGTACTGGTCGAATGG | OsAOS2 定量 PCR |
| OsAOS2-QR | AAGGTGTCGTACCGGAGGAA | qRT-PCR of OsAOS2 |
| OsPAL1-QF | AGGAGCTCGGCTGCGTATT | OsPAL1 定量 PCR |
| OsPAL1-QR | ATGCCGAGGAACACCTTGTT | qRT-PCR of OsPAL1 |
| OsAOC-QF | CCAAGGTGCAGGAGATGTT | OsAOC 定量 PCR |
| OsAOC-QR | TACAGCTTGTTGGTGAAGGG | qRT-PCR of OsAOC |
| OsPBZ1-QF | CCCTGCCGAATACGCCTAA | OsPBZ1 定量 PCR |
| OsPBZ1-QR | CTCAAACGCCACGAGAATTTG | qRT-PCR of OsPBZ1 |
表1 本研究用到的引物列表
Table 1. Primers used in this study
| 引物名称 Primer name | 序列(5’→3’) Primer sequence(5’→3’) | 用途 Purpose |
|---|---|---|
| OsOPR10-F | ATGAAGTCAAGTCCAAATCAGCTG | CDS扩增 |
| OsOPR10-R | ACGAGTTGCTAGTTCTGAATTCTGAT | CDS amplification |
| OsOPR10-OE-F | CTTCTGCAGCCCGGGGGATCCATGAAGTCAAGTCCAAATCAGCTG | 过表达载体构建 |
| OsOPR10-OE-R | CGATCGGGGAAATTCGGATCCTCAAAGATCTTCTTCAGAAATCAACTTTTGTTCACGAGTTGCTAGTTCTGAATTCTGAT | Construction of overexpression vector |
| UbiP-seq | TTTTAGCCCTGCCTTCATACGC | 过表达载体测序 |
| NosR-seq | AGACCGGCAACAGGATTCAATC | Overexpression vector sequencing |
| OsOPR10-gRT1 | CGGCCGCCTGCTTCCGTTCGTTTTAGAGCTAGAAAT | CRISPR/Cas9第一个靶点扩增 |
| OsOPR10-OsU3T1 | GAACGGAAGCAGGCGGCCGTGCCACGGATCATCTGC | Amplification of the first CRISPR/Cas9 target |
| OsOPR10-gRT2 | AGATCGGCGCGCACCGCCTGTTTTAGAGCTAGAAAT | CRISPR/Cas9第二个靶点扩增 |
| OsOPR10-U6aT2 | AGGCGGTGCGCGCCGATCTCGGCAGCCAAGCCAGCA | Amplification of the second CRISPR/Cas9 target |
| OsOPR10-T1-F | AGAGAACCGGTGCCGCTTC | CRISPR/Cas9第一个靶点验证 |
| OsOPR10-T1-R | GGTCGGAGTCGTGGCAGTC | First CRISPR/Cas9 target validation |
| Actin-QF | GAGTATGATGAGTCGGGTCCAG | Actin 定量 PCR |
| Actin-QR | ACACCAACAATCCCAAACAGAG | qRT-PCR of Actin |
| OsUG-F | TTCTGGTCCTTCCACTTTCAG | 泛素融合蛋白定量PCR |
| OsUG-R | ACGATTGATTTAACCAGTCCATGA | qRT-PCR of ubiquitin fusion protein OsUG |
| MoPot2-F | ACGACCCGTCTTTACTTATTTGG | MoPot2 定量 PCR |
| MoPot2-R | AAGTAGCGTTGGTTTTGTTGGAT | qRT-PCR of MoPot2 |
| OsOPR10-BD-F | TCAGAGGAGGACCTGCATATGATGAAGTCAAGTCCAAATCAGCTG | OsOPR10-BD载体构建 |
| OsOPR10-BD-R | TCGACGGATCCCCGGGAATTCTCAACGAGTTGCTAGTTCTGAATTC | Construction of OsOPR10-BD vector |
| OsCYP28-AD-F | GTACCAGATTACGCTCATATGATGGTGTTGCCTTCATCAAA | OsCYP28-AD载体构建 |
| OsCYP28-AD-R | ATGCCCACCCGGGTGGAATTCTTATGGCAGACTTGGAGGGG | Construction of OsCYP28-AD vector |
| OsOPR10-pGDG-F | gcttcgaattctgcagtcgacATGAAGTCAAGTCCAAATCAGCTG | OsOPR10-pGDG载体构建 |
| OsOPR10-pGDG-R | ttatctagatccggtggatccTCAACGAGTTGCTAGTTCTGAATTC | Construction of OsOPR10-pGDG vector |
| OsOPR10-PHF223-F | CGGGATCGATGAAGTCAAGTCCAAATCAG | OsOPR10-PHF223载体构建 |
| OsOPR10-PHF223-R | GGGGTACCACGAGTTGCTAGTTCTGAATT | Construction of OsOPR10-PHF223 vector |
| OsPR1a-QF | CGTCTTCATCACCTGCAACTACTC | OsPR1a 定量 PCR |
| OsPR1a-QR | CATGCATAAACACGTAGCATAGCA | qRT-PCR of OsPR1a |
| OsAOS2-QF | CAATACGTGTACTGGTCGAATGG | OsAOS2 定量 PCR |
| OsAOS2-QR | AAGGTGTCGTACCGGAGGAA | qRT-PCR of OsAOS2 |
| OsPAL1-QF | AGGAGCTCGGCTGCGTATT | OsPAL1 定量 PCR |
| OsPAL1-QR | ATGCCGAGGAACACCTTGTT | qRT-PCR of OsPAL1 |
| OsAOC-QF | CCAAGGTGCAGGAGATGTT | OsAOC 定量 PCR |
| OsAOC-QR | TACAGCTTGTTGGTGAAGGG | qRT-PCR of OsAOC |
| OsPBZ1-QF | CCCTGCCGAATACGCCTAA | OsPBZ1 定量 PCR |
| OsPBZ1-QR | CTCAAACGCCACGAGAATTTG | qRT-PCR of OsPBZ1 |

图1 外源施加MeJA(A)和SA(B)后OsOPR10基因的表达模式 图中数据为平均值±标准差,*、**和***分别代表处理间在 P<0.05,P<0.01和P<0.001水平上差异显著(t-检验)。下图同。
Fig. 1. Expression patterns of OsOPR10 after MeJA (A) and SA (B) application Data in the figure are Mean±SD, and *, ** and ***represent significant difference at the 0.05, 0.01 and 0.001 levels (t-test), respectively. The same below.

图3 OsOPR10基因敲除靶点设计和突变类型鉴定 A: OsOPR10的2个gRNA靶点;B: OsOPR10敲除植株的突变位点检测。
Fig. 3. Knockout target design and mutation type identification of OsOPR10 A, Two gRNA targets of OsOPR10 gene; B, Detection of mutantion sites in OsOPR10 knockout transgenic line.

图5 OsOPR10转基因水稻对稻瘟病的抗性分析 A: 造伤接种稻瘟病菌后的发病情况;B: 造伤接种稻瘟病菌后的发病面积;C: 造伤接种稻瘟病菌后的相对真菌生物量,图中数据为平均值±标准差, ***分别代表处理间在 P<0.001 水平上差异达显著水平(t-检验), n=4。
Fig. 5. Response of OsOPR10 transgenic rice to M. oryzae A, Lesions after inoculation with M. oryzae; B, Lesion area after inoculation with M. oryzae; C, Relative fungal biomass after inoculation with M. oryzae. Data in the figure are Mean±SD, and *** represent significant difference at 0.001 level(t-test), respectively, n=4.

图6 OsOPR10转基因水稻对白叶枯菌的抗性分析 A:剪叶接种白叶枯菌后的发病情况;B:接种白叶枯菌后的病斑长度(平均值±标准差),* P<0.05 (t-检验), n=5。
Fig. 6. Response of OsOPR10 transgenic rice to Xoo. A, Lesions size after inoculation by Xoo by leaf clipping method; B, Length of lesions after inoculation with Xoo. Data in the figure are Mean±SD, and * represents significant difference at the 0.05 levels (t-test), n=5.

图7 OsOPR10转基因水稻对几丁质和flg22诱导的响应 A: OsOPR10-OE转基因水稻和日本晴经几丁质诱导后活性氧爆发情况; B: OsOPR10-KO转基因水稻和日本晴经几丁质诱导后活性氧爆发情况; C: OsOPR10-OE转基因水稻和日本晴经flg22诱导后活性氧爆发情况; D: OsOPR10-KO转基因水稻和日本晴经flg22诱导后活性氧爆发情况。
Fig. 7. Response of OsOPR10 transgenic rice to chitin and flg22 induction. A, Chitin-induced ROS burst in OsOPR10-OE transgenic rice and Nipponbare; B, Chitin-induced ROS burst in OsOPR10-KO transgenic rice and Nipponbare; C, flg22-induced ROS burst in OsOPR10-OE transgenic rice and Nipponbare; D, flg22-induced ROS burst in OsOPR10-KO transgenic rice and Nipponbare.
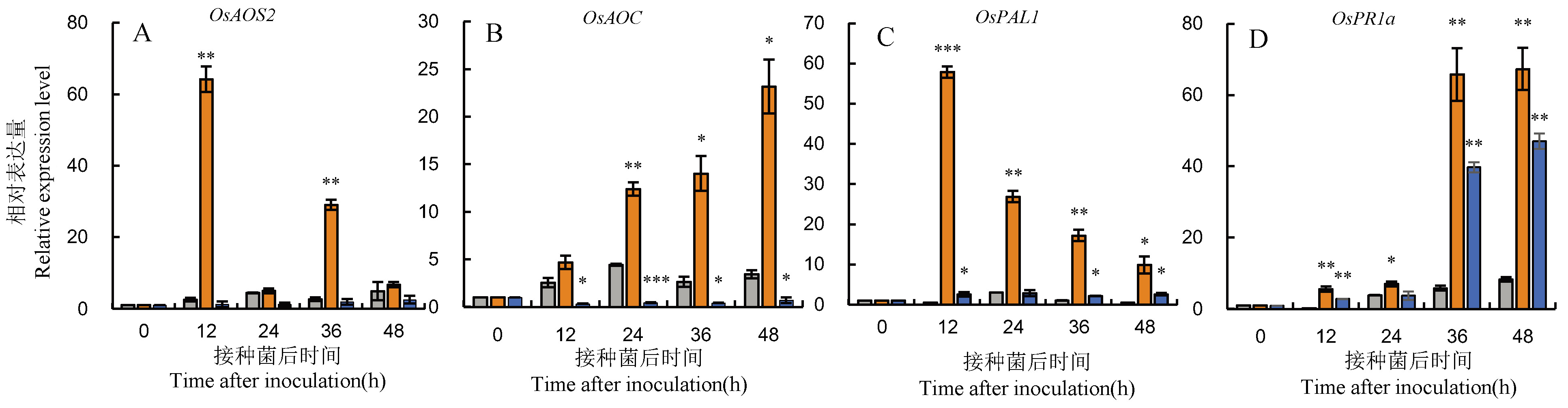
图8 OsOPR10转基因水稻在稻瘟菌侵染后JA和SA途径基因的表达分析 图中数据为平均值±标准差,*、**和***分别代表处理间在 P<0.05、P<0.01 和 P<0.001水平上差异达显著水平(t-检验)。
Fig. 8. Expression analysis of genes involved in JA and SA pathways in OsOPR10 transgenic rice after infection by M. oryzae Data in the figure are Mean±SD, *, ** and *** represent significant difference at the 0.05, 0.01 and 0.001 levels (t-test), respectively.

图9 OsOPR10-PHF223和OsOPR10-pGDG融合蛋白的亚细胞定位 A: OsOPR10-PHF223融合蛋白和PHF223载体在水稻原生质体中的亚细胞定位,图中标尺表示10 μm; B: OsOPR10-pGDG融合蛋白和pGDG载体在烟草叶片中的亚细胞定位,图中标尺表示25 μm。
Fig. 9. Subcellular localization of OsOPR10-PHF223 and OsOPR10-pGDG fusion protein A, Subcellular localization of OsOPR10-PHF223 fusion protein and PHF223 vector in rice protoplasts, the scale bar in the figure represents 10 μm; B, Subcellular localization of OsOPR10-pGDG fusion protein and pGDG vector in tobacco leaves, the scale bar in the figure represents 25 μm.

图11 荧光素酶互补试验验证OsOPR10和OsCYP28互作
Fig. 11. Luciferase complementation assay verifies the interaction between OsOPR10 and OsCYP28 a: OsOPR10-Nluc+OsCYP28-Cluc; b: OsOPR10-Nluc+Cluc; c: OsCYP28-Cluc+Nluc; d: Nluc+Cluc.
| [1] | Ainsworth E A. Rice production in a changing climate: A meta-analysis of responses to elevated carbon dioxide and elevated ozone concentration[J]. Global Change Biology, 2010, 14(7): 1642-1650. |
| [2] | Dean R, Kan J, Pretorius Z A, Hammond-Kosack K E, Pietro A D, Spanu P D, Ruddd J J, Pickman M. The Top 10 fungal pathogens in molecular plant pathology[J]. Molecular Plant Pathology, 2012, 13(4): 414-430. |
| [3] | Jiang N, Yan J, Liang Y, Shi Y L, He Z Z, Wu Y T, Zeng Q, Liu X L, Peng J H. Resistance genes and their interactions with bacterial blight pathogens in rice[J]. Rice, 2020, 13(1): 1-12. |
| [4] | Liu W D, Wang G L. Plant innate immunity in rice: A defense against pathogen infection[J]. National Science Review, 2016, 3: 295-308. |
| [5] | Yuan M H, Jiang Z Y, Bi G Z, Nomura K Y, Liu M H, Wang Y P, Cai B Y, Zhou J M, He S Y, Xin X F. Pattern- recognition receptors are required for NLR-mediated plant immunity[J]. Nature, 2021, 592(7852): 105-109. |
| [6] | 覃磊, 彭志红, 夏石头. 植物NLR免疫受体的识别、免疫激活与信号调控[J]. 植物学报, 2022, 57(1): 12-23. |
| Qin L, Peng Z H, Xia S T. Recognition, immune activation and signal regulation of plant NLR immune receptor[J]. Chinese Bulletin of Botany, 2022, 57(1): 12-23. (in Chinese with English abstract) | |
| [7] | Ramírez-Zavaleta C Y, García-Barrera L J, Rodríguez- Verástegui L L, Arrieta-Flores D. An overview of PRR-and NLR-mediated immunities: Conserved signaling components across the plant kingdom that communicate both pathways[J]. International Journal of Molecular Sciences, 2022, 23(21): 12974. |
| [8] | Qiu J, Xie J, Chen Y, Shen Z, Shi H, Naqvi N I, Qian Q, Liang Y, Kou Y J. Warm temperature compromises JA-regulated basal resistance to enhance Magnaporthe oryzae infection in rice[J]. Molecular Plant, 2022, 15(4): 723-739. |
| [9] | Wasternack C, Hause B. Jasmonates and octadecanoids: Signals in plant stress responses and development[J]. Progress in Nucleic Acid Research and Molecular Biology, 2002, 72: 165-221. |
| [10] | Liechti R. The jasmonate pathway[J]. Science, 2002, 296(5573): 1649-1650. |
| [11] | Al-Momany B, Abu-Romman S. Homologs of old yellow enzyme in plants[J]. Australian Journal of Crop Science. 2016, 10(4): 584-590. |
| [12] | Agrawal G K, Jwa N S, Shibato J, Han O, Iwahashi H, Rakwal R. Diverse environmental cues transiently regulate OsOPR1 of the octadecanoid pathway revealing its importance in rice defense/stress and development[J]. Biochemical and Biophysical Research Communications, 2003, 310(4): 1073-1082. |
| [13] | Guo H M, Li H C, Zhou S R, Xue H W, Miao X X. Cis-12-oxo-phytodienoic acid stimulates rice defense response to a piercing-sucking insect[J]. Molecular Plant, 2014, 7(11): 1683-1692. |
| [14] | Tani T, Sobajima H, Okada K, Chujo T, Arimura S, Tsutsumi N, Nishimura M, Seto H, Nojiri H, Yamane H. Identification of OsOPR7 encoding 12-oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice: An international journal of plant biology[J]. Planta, 2008, 227(3): 517-526. |
| [15] | Cong L, Ran F A, Cox D, Lin S L, Barretto R, Habib N, Hsu P D, Wu X, Jiang W Y, Marraffini L A, Zhang F. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823. |
| [16] | Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V. Editing plant genomes with CRISPR/ Cas9[J]. Current Opinion in Biotechnology, 2015, 32: 76-84. |
| [17] | 曾栋昌, 马兴亮, 谢先荣, 祝钦泷, 刘耀光. 植物CRISPR/Cas9多基因编辑载体构建和突变分析的操作方法[J]. 中国科学: 生命科学, 2018, 48(7): 12. |
| Zeng D C, Ma X L, Xie X R, Zhu Q L, Liu Y G. Operational methods for the construction of plant CRISPR/Cas9 multi-gene editing vectors and mutation analysis[J]. Science China: Life Sciences, 2018, 48(7): 12. (in Chinese with English abstract) | |
| [18] | Clarke J D. Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation[J]. Cold Spring Harbor Protocol, 2009(3): pdb.prot5177. |
| [19] | Shi X T, Long Y, He F, Zhang C Y, Wang R Y, Zhang T, Wu W, Hao Z Y, Wang Y, Wang G L, Ning Y S. The fungal pathogen Magnaporthe oryzae suppresses innate immunity by modulating a host potassium channel[J]. PLoS Pathogens, 2018, 14(1): e1006878. |
| [20] | Hao Z Y, Tian J F, Fang H, Fang L, Xu X, He F, Li S Y, Xie W Y, Du Q, You X M, Wang D B, Chen Q H, Wang R Y, Zuo S M. A VQ-motif-containing protein fine-tunes rice immunity and growth by a hierarchical regulatory mechanism[J]. Cell Reports, 2022, 40(7): 111235. |
| [21] | Park C H, Chen S, Shirsekar G, Zhou B, Khang C H, Songkumarn P, Afzal A J, Ning Y S, Wang R Y, Bellizzi M, Valent B, Wang G L. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice[J]. Plant Cell, 2012, 24(11): 4748-4762. |
| [22] | Yoo S D, Cho Y H, Sheen J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis[J]. Nature Protocols, 2007, 2(7): 1565-1572. |
| [23] | Li R L, Wang J L, Xu L, Sun M H, Yi K K, Zhao H Y. Functional analysis of phosphate transporter OsPHT4 family members[J]. Rice Science, 2020, 27(6): 493-503. |
| [24] | Gu Y H, Li G N, Wang P, Guo Y, Li J R. A simple and precise method (Y2H-in-frame-seq) improves yeast two-hybrid screening with cDNA libraries[J]. Journal of Genetics Genomics, 2022, 49(6): 595-598. |
| [25] | Mou Y F, Liu Y Y, Tian S J, Guo Q P, Wang C S, Wen S S. Genome-Wide identification and characterization of the OPR gene family in wheat[J]. International Journal of Molecular Sciences, 2019, 20(8): 1914. |
| [26] | Zhang J L, Simmons C, Yalpani N, Crane V, Wilkinson H, Kolomiets M. Genomic analysis of the 12-oxo- phytodienoic acid reductase gene family of Zea mays[J]. Plant Molecular Biology, 2005, 59: 323-334. |
| [27] | Piñera-Chavez F J, Berry P M, Foulkes M J, Molero G, Reynolds M P. Avoiding lodging in irrigated spring wheat. II. Genetic variation of stem and root structural properties[J]. Field Crops Research, 2016, 196: 64-74. |
| [28] | Scalschi L, Sanmartín M, Camañes G, Troncho P, Sánchez-Serrano J J, García-Agustín P, Vicedo B. Silencing of OPR3 in tomato reveals the role of OPDA in callose deposition during the activation of defense responses against Botrytis cinerea[J]. Plant Journal, 2015, 81(2): 304-315. |
| [29] | Li C, Xu M X, Cai X, Han Z G, Si J P, Chen D H. Jasmonate signaling pathway modulates plant defense, growth, and their trade-offs[J]. International Journal of Molecular Sciences, 2022, 23(7): 3945. |
| [30] | 文锴, 王远, 胡蝶, 袁敬平, 侯喜林, 李英. 不结球白菜BcOPR3基因的克隆与功能分析[J]. 南京农业大学学报, 2017, 40(5): 8. |
| Wen K, Wang Y, Hu D, Yuan J P, Hou X L, Li Y. Cloning and expression analysis of BcOPR3 gene in non-heading Chinese cabbage[J]. Journal of Nanjing Agricultural University, 2017, 40(5): 8. (in Chinese with English abstract) | |
| [31] | 王艳微, 王敏, 王江, 张庆祝, 解莉楠. 大豆OPR基因家族全基因组鉴定与表达分析[J]. 大豆科学, 2022, 41(2): 129-139. |
| Wang Y W, Wang M, Wang J, Zhang Q Z, Xie L N. Genome-wide identification and expression analysis of soybean OPR gene family[J]. Soybean Science, 2022, 41(2): 129-139. (in Chinese with English abstract) | |
| [32] | Dong W, Wang M C, Xu F, Quan T Y, Peng K Q, Xiao L T, Xia G M. Wheat oxophytodienoate reductase gene TaOPR1 confers salinity tolerance via enhancement of abscisic acid signaling[J]. Plant Physiology, 2013, 161(3): 1217-1228. |
| [33] | Wang Y K, Yuan G L, Yuan S H, Duan W J, Wang P, Bai J F, Zhang F T, Gao S Q, Zhang L P, Zhao C P. TaOPR2 encodes a 12-oxo-phytodienoic acid reductase involved in the biosynthesis of jasmonic acid in wheat (Triticum aestivum L.)[J]. Biochemical and Biophysical Research Communications, 2016, 470(1): 233-238. |
| [34] | 游双红, 谭平, 史文景, 武峥, 陈元平, 伊洪伟, 周广文. 葡萄OPR家族全基因组鉴定及重金属胁迫下的表达分析[J]. 基因组学与应用生物学, 2021, 40(5): 10. |
| You S H, Tan P, Shi W J, Wu Z, Chen Y P, Yi H W, Zhou G W. Genome-wide identification of OPR family genes and the expression of these genes in response to heavy metal stress[J]. Genomics and Applied Biology, 2021, 40(5): 10. (in Chinese with English abstract) | |
| [35] | Li M P, Kim C H. Chloroplast ROS and stress signaling[J]. Plant Communication, 2021, 3(1): 100264. |
| [36] | Gao H X, Zhu L, Liu T Q, Leng X Y, Zhu Z X, Xie W, Lü H T, Jin Z X, Wu P, Zhang Z C. Identification of a novel OsCYP2 allele that was involved in rice response to low temperature stress[J]. Phyton-International Journal of Experimental Botany, 2023, 92(6): 1743-1763. |
| [37] | Zhu W N, Xu L Q, Yu X X, Zhong Y. The immunophilin CYCLOPHILIN28 affects PSII-LHCII supercomplex assembly and accumulation in Arabidopsis thaliana[J]. Journal of Integrative Plant Biology, 2022, 64(4): 915-929. |
| [1] | 汪邑晨, 朱本顺, 周磊, 朱骏, 杨仲南. 光/温敏核不育系的不育机理及两系杂交稻的发展与展望 [J]. 中国水稻科学, 2024, 38(5): 463-474. |
| [2] | 许用强, 徐军, 奉保华, 肖晶晶, 王丹英, 曾宇翔, 符冠富. 水稻花粉管生长及其对非生物逆境胁迫的响应机理研究进展 [J]. 中国水稻科学, 2024, 38(5): 495-506. |
| [3] | 何勇, 刘耀威, 熊翔, 祝丹晨, 王爱群, 马拉娜, 王廷宝, 张健, 李建雄, 田志宏. 利用CRISPR/Cas9技术编辑OsOFP30基因创制水稻粒型突变体 [J]. 中国水稻科学, 2024, 38(5): 507-515. |
| [4] | 吕阳, 刘聪聪, 杨龙波, 曹兴岚, 王月影, 童毅, Mohamed Hazman, 钱前, 商连光, 郭龙彪. 全基因组关联分析(GWAS)鉴定水稻氮素利用效率候选基因 [J]. 中国水稻科学, 2024, 38(5): 516-524. |
| [5] | 杨好, 黄衍焱, 王剑, 易春霖, 石军, 谭楮湉, 任文芮, 王文明. 水稻中八个稻瘟病抗性基因特异分子标记的开发及应用 [J]. 中国水稻科学, 2024, 38(5): 525-534. |
| [6] | 杨铭榆, 陈志诚, 潘美清, 张汴泓, 潘睿欣, 尤林东, 陈晓艳, 唐莉娜, 黄锦文. 烟-稻轮作下减氮配施生物炭对水稻茎鞘同化物转运和产量 形成的影响 [J]. 中国水稻科学, 2024, 38(5): 555-566. |
| [7] | 熊家欢, 张义凯, 向镜, 陈惠哲, 徐一成, 王亚梁, 王志刚, 姚坚, 张玉屏. 覆膜稻田施用炭基肥对水稻产量及氮素利用的影响 [J]. 中国水稻科学, 2024, 38(5): 567-576. |
| [8] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [9] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [10] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [11] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [12] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [13] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [14] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [15] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||