
中国水稻科学 ›› 2023, Vol. 37 ›› Issue (4): 359-367.DOI: 10.16819/j.1001-7216.2023.221113
李刚1, 高清松2, 李伟2, 张雯霞2, 王健1, 程保山1, 王迪1, 高浩1, 徐卫军1, 陈红旗3( ), 纪剑辉2(
), 纪剑辉2( )
)
收稿日期:2022-11-24
修回日期:2023-02-08
出版日期:2023-07-10
发布日期:2023-07-17
通讯作者:
*email: chqhzfy@126.com;jijianhui@hytc.edu.cn
基金资助:
LI Gang1, GAO Qingsong2, LI Wei2, ZHANG Wenxia2, WANG Jian1, CHEN Baoshan1, WANG Di1, GAO Hao1, XU Weijun1, CHEN Hongqi3( ), JI Jianhui2(
), JI Jianhui2( )
)
Received:2022-11-24
Revised:2023-02-08
Online:2023-07-10
Published:2023-07-17
Contact:
*email: chqhzfy@126.com;jijianhui@hytc.edu.cn
摘要:
【目的】 为改良高产粳稻品种淮119株高偏高及易感稻瘟病等不利性状,利用CRISPR/Cas9基因编辑技术对淮119中SD1基因进行定向敲除,为淮119后代品种改良奠定基础。【方法】 利用CRISPR/Cas9系统,以SD1基因为靶基因,构建基因敲除载体,以农杆菌介导转化淮119,获得无转基因插入的纯合突变株,进一步对纯合突变株株高、农艺性状、稻瘟病抗性及氮素吸收利用能力等进行综合分析。【结果】 利用农杆菌转化淮119,鉴定获得1株无转基因载体序列插入的纯合突变株。大田种植发现,野生型淮119出现60%面积以上的倒伏,而sd1纯合突变株群体由于株高变矮,从而有效避免了生育后期的倒伏。此外,用不同浓度的GA(0.01~1.00 μmol/L)处理,野生型苗高均显著高于突变株,表明sd1突变导致对外源GA敏感性降低。稻瘟病抗性鉴定结果表明,sd1基因突变不仅有效降低株高,同时增强了对稻瘟病的抗性水平。不利因素是,SD1基因的敲除降低了淮119的氮素吸收利用效率。【结论】 淮119中SD1基因定向敲除获得的无转基因插入纯合突变株,不仅株高显著下降,且稻瘟病抗性得到增强。
李刚, 高清松, 李伟, 张雯霞, 王健, 程保山, 王迪, 高浩, 徐卫军, 陈红旗, 纪剑辉. 定向敲除SD1基因提高水稻的抗倒性和稻瘟病抗性[J]. 中国水稻科学, 2023, 37(4): 359-367.
LI Gang, GAO Qingsong, LI Wei, ZHANG Wenxia, WANG Jian, CHEN Baoshan, WANG Di, GAO Hao, XU Weijun, CHEN Hongqi, JI Jianhui. Directed Knockout of SD1 Gene Improves Lodging Resistance and Blast Resistance of Rice[J]. Chinese Journal OF Rice Science, 2023, 37(4): 359-367.
| 引物名称 Primer name | 序列(5’-3’) Primer sequence(5'-3') |
|---|---|
| SD1-P1 | TGTGGAGCCATTCGTGTGGCCGAA |
| SD1-P2 | AAACTTCGGCCACACGAATGGCTC |
| SD1-F | CACAGCCGCACCAACCAC |
| SD1-R | GTGGAAGCCGAAGGAGAGG |
| HPTF | GGGTGTCACGTTGCAAGACC |
| HPTR | ATGCCTCCGCTCGAAGTAGC |
| sgRNA-F | TCCCAGTCACGACGTTGTAA |
| sgRNA-R | GGCCATTTGTCTGCAGAAT |
| Cas9-F | CACCATCTACCACCTGAGAA |
| Cas9-R | CGAAGTTGCTCTTGAAGTTG |
| Actin-F | CCCCTCCTGAAAGGAAGTA |
| Actin-R | GGTCCGAAGAATTAGAAGCA |
表1 本研究中使用的引物序列
Table 1. Primer sequences used in this study.
| 引物名称 Primer name | 序列(5’-3’) Primer sequence(5'-3') |
|---|---|
| SD1-P1 | TGTGGAGCCATTCGTGTGGCCGAA |
| SD1-P2 | AAACTTCGGCCACACGAATGGCTC |
| SD1-F | CACAGCCGCACCAACCAC |
| SD1-R | GTGGAAGCCGAAGGAGAGG |
| HPTF | GGGTGTCACGTTGCAAGACC |
| HPTR | ATGCCTCCGCTCGAAGTAGC |
| sgRNA-F | TCCCAGTCACGACGTTGTAA |
| sgRNA-R | GGCCATTTGTCTGCAGAAT |
| Cas9-F | CACCATCTACCACCTGAGAA |
| Cas9-R | CGAAGTTGCTCTTGAAGTTG |
| Actin-F | CCCCTCCTGAAAGGAAGTA |
| Actin-R | GGTCCGAAGAATTAGAAGCA |
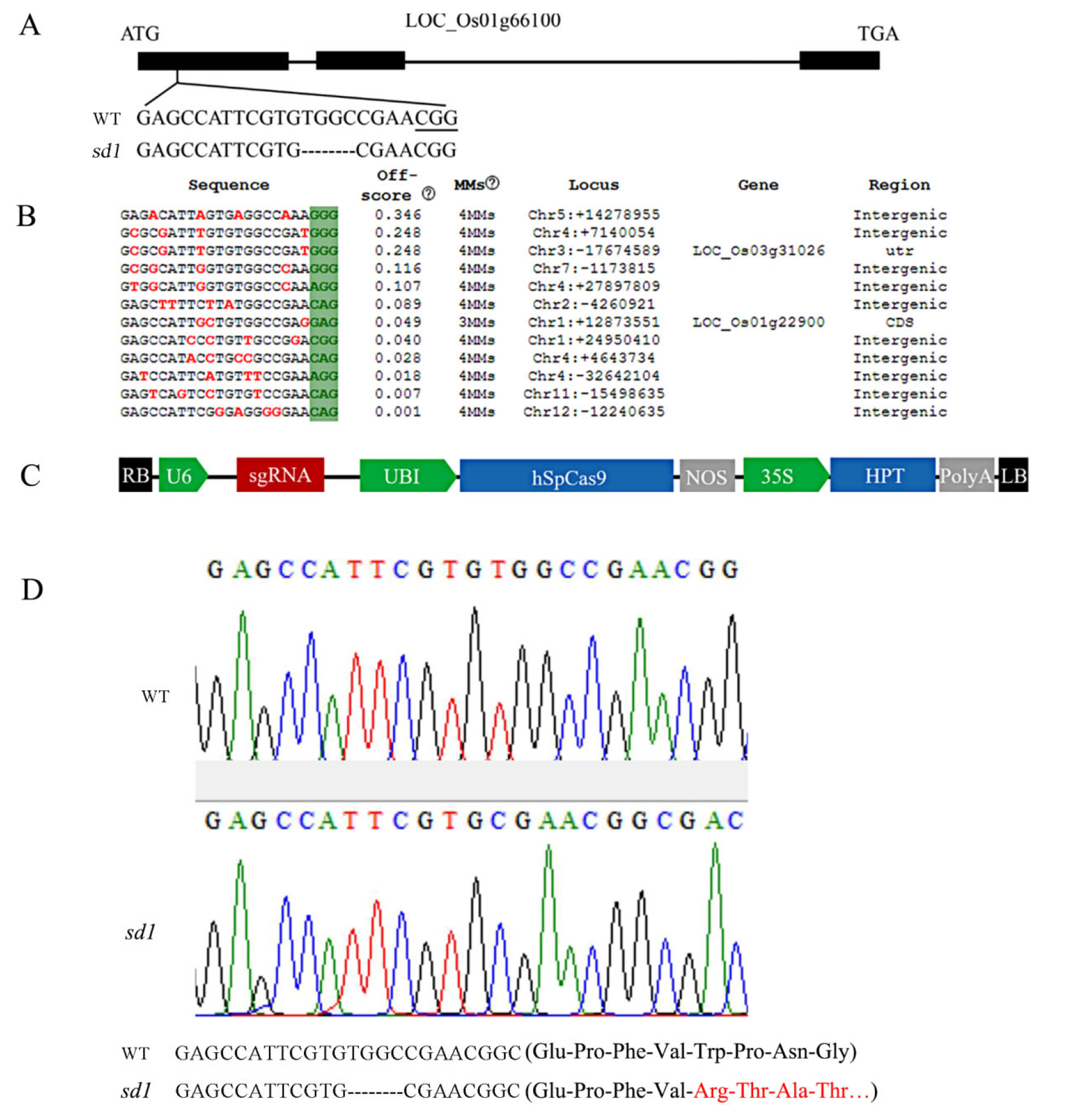
图1 SD1基因敲除靶点设计、T-DNA结构和突变鉴定 A―SD1基因结构及基因敲除靶点;B―脱靶序列分析;C―基因编辑载体T-DNA区段结构;D―测序结果及相应氨基酸序列。
Fig. 1. Target site for SD1 gene knock-out design, T-DNA structure and mutation identification. A, SD1 gene structure and gene knockout target sites; B, Off target sequence analysis; C, Gene editing vector T-DNA segment structure; D, Sequencing results and corresponding amino acid sequence.
| 试验小区 Experiment plot | 倒伏比例 Lodging percentage/% | Yield 产量/(t·hm−2) | ||
|---|---|---|---|---|
| 野生型 Wild type | sd1 | 野生型 Wild type | sd1 | |
| 重复小区1 Repeat 1 | 60 | 0 | 10.52 | 10.31 |
| 重复小区2 Repeat 2 | 90 | 0 | 11.26 | 11.53 |
| 重复小区3 Repeat 3 | 75 | 0 | 11.55 | 11.46 |
| 平均 Mean | 75 | 0 | 11.11 | 11.10 |
表2 野生型淮119及sd1突变体群体产量及其倒伏比例
Table 2. Yield and lodging result of the wild-type Huai 119 and sd1 mutant.
| 试验小区 Experiment plot | 倒伏比例 Lodging percentage/% | Yield 产量/(t·hm−2) | ||
|---|---|---|---|---|
| 野生型 Wild type | sd1 | 野生型 Wild type | sd1 | |
| 重复小区1 Repeat 1 | 60 | 0 | 10.52 | 10.31 |
| 重复小区2 Repeat 2 | 90 | 0 | 11.26 | 11.53 |
| 重复小区3 Repeat 3 | 75 | 0 | 11.55 | 11.46 |
| 平均 Mean | 75 | 0 | 11.11 | 11.10 |
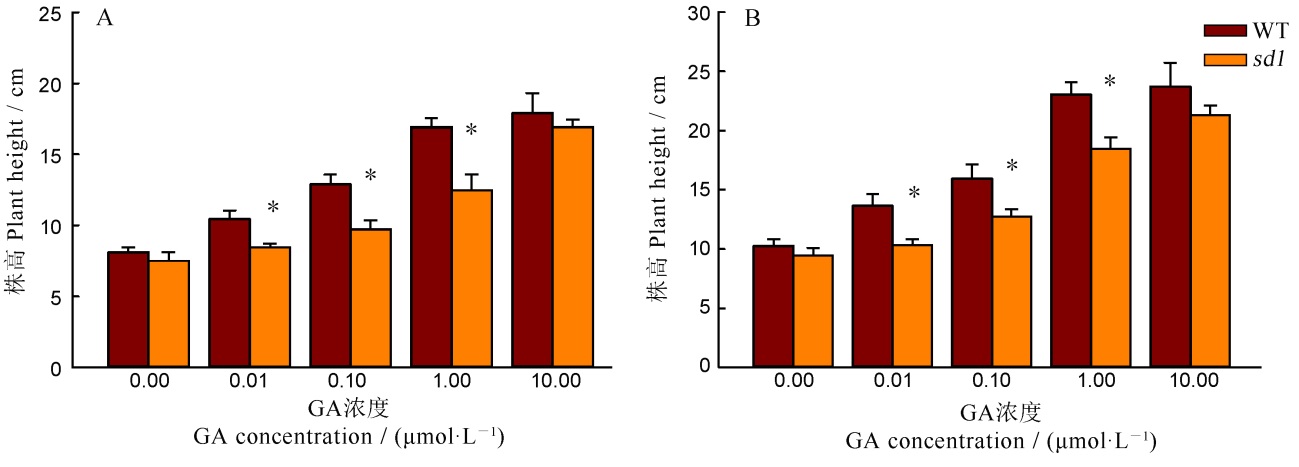
图4 外源GA处理下野生型与突变体株高比较 A—GA处理7 d; B—GA处理10 d. *表示突变体与野生型间的差异达0.05显著水平(n=4)。
Fig. 4. Plant height of the wild type and the mutant under the exogenous GA treatment. A, GA treatment for 7 days; B, GA treatment for 10 days; *Significant difference at 0.05 level between the wild type and the mutant (n=4).
| 品种名称Variety name | 苗瘟病级Disease rating of seedling blast | 叶瘟病级Disease rating of leaf blast | 穗颈瘟Neck blast | 抗性 Resistance | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 发病率Incidence rate of blast/% | 病级Disease rating | 损失率 Loss rate of blast /% | 病级 Disease rating | 抗性综合指数Comprehensive resistance index | 病级Disease rating | |||||
| sd1 | 0 | 1 | 23 | 5 | 6 | 3 | 3.00 | 3 | 中抗Moderately resistant | |
| 淮119 Huai 119 | 0 | 2 | 82 | 9 | 33 | 7 | 6.25 | 7 | 感病Sensitive | |
表3 sd1及淮119对水稻不同时期稻瘟病的抗性鉴定结果
Table 3. Resistance evaluation of sd1 and Huai 119 to rice blast.
| 品种名称Variety name | 苗瘟病级Disease rating of seedling blast | 叶瘟病级Disease rating of leaf blast | 穗颈瘟Neck blast | 抗性 Resistance | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 发病率Incidence rate of blast/% | 病级Disease rating | 损失率 Loss rate of blast /% | 病级 Disease rating | 抗性综合指数Comprehensive resistance index | 病级Disease rating | |||||
| sd1 | 0 | 1 | 23 | 5 | 6 | 3 | 3.00 | 3 | 中抗Moderately resistant | |
| 淮119 Huai 119 | 0 | 2 | 82 | 9 | 33 | 7 | 6.25 | 7 | 感病Sensitive | |
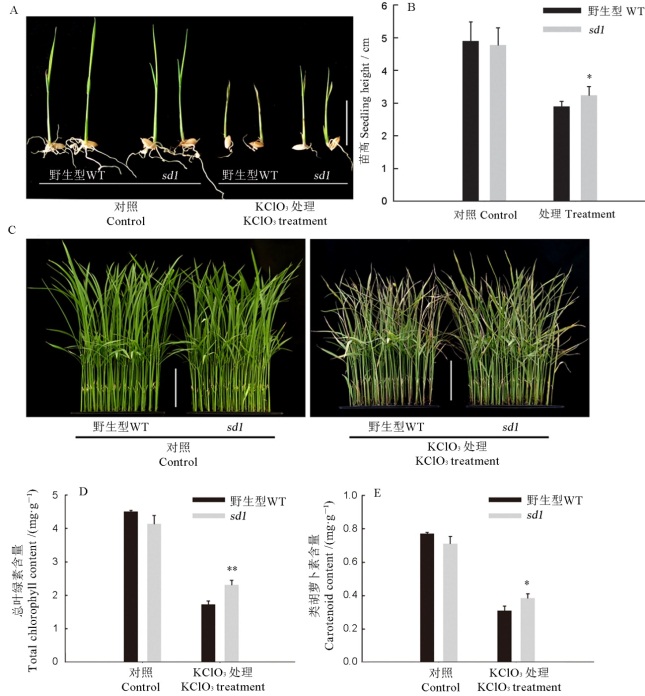
图6 野生型淮119和sd1突变体氯酸盐处理下的表型 A―萌发期KClO3处理的幼苗,比例尺=2 cm;B―萌发期KClO3处理的苗高,n=10;C―苗期KClO3处理的幼苗,比例尺=5 cm;D和E―苗期氯酸盐处理后幼苗的总叶绿素(D)和类胡萝卜素(E)含量。*P<0.05; ***P<0.01.
Fig. 6. Phenotype of the wild type Huai 119 and sd1 mutant under the chlorate treatment. A, Seedlings treated with KClO3 during the germination period, Bar= 2 cm; B, Height of seedlings treated with KClO3 during germination, n=10; C, Seedlings treated with KClO3 at seedling stage, scale=5 cm; D and E, Total chlorophyll (D) and carotenoid (E) contents of seedlings after KClO3 treatment at seedling stage.
| [1] | Doebley J F, Gaut B S, Smith B D. The molecular genetics of crop domestication[J]. Cell, 2006, 127(7): 1309-1321. |
| [2] | Cho S W, Kim S, Kim J M, Kim J S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease[J]. Nature Biotechnology, 2013, 31(3): 230-232. |
| [3] | Cong L, Ran F A, Cox D, Lin S L, Barretto R, Habib N, Hsu P D, Wu X B, Jiang W Y, Marraffini L A, Zhang F. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823. |
| [4] | Jinek M, East A, Cheng A R, Lin S, Ma E B, Doudna J. RNA-programmed genome editing in human cells[J]. Elife, 2013, 2: e00471. |
| [5] | Mali P, Aach J, Stranges P B, Esvelt K M, Moosburner M, Kosuri S, Yang L H, Church G M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering[J]. Nature Biotechnology, 2013, 31(9): 833-838. |
| [6] | Li Y, Li W J, Li J. The CRISPR/Cas9 revolution continues: From base editing to prime editing in plant science[J]. Journal of Genetics and Genomics, 2021, 48(8): 661-670. |
| [7] | Chen K L, Wang Y P, Zhang R, Zhang H W, Gao C X. CRISPR/Cas genome editing and precision plant breeding in agriculture[J]. Annual Review of Plant Biology, 2019, 70: 667-697. |
| [8] | 沈兰, 李健, 付亚萍, 王俊杰, 华宇峰, 焦晓真, 严长杰, 王克剑. 利用CRISPR/Cas9系统定向改良水稻粒长和穗粒数性状[J]. 中国水稻科学, 2017, 31(3): 223-231. |
| Shen L, Li J, Fu Y P, Wang J J, Hua Y F, Jiao X Z, Yan C J, Wang K J. Orientation improvement of grain length and grain number in rice by using CRISPR/Cas9 system[J]. Chinese Journal of Rice Science, 2017, 31(3): 223-231. (in Chinese with English abstract) | |
| [9] | Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush G S, Kitano H, Matsuoka M. Green revolution: A mutant gibberellin-synthesis gene in rice[J]. Nature, 2002, 416(6882): 701-702. |
| [10] | Peng Y L, Hu Y G, Qian Q, Ren D Y. Progress and prospect of breeding utilization of green revolution gene SD1 in rice[J]. Agriculture, 2021, 11(7): 611. |
| [11] | Spielmeyer W, Ellis M H, Chandler P M. Semidwarf(sd-1), "green revolution" rice, contains a defective gibberellin 20-oxidase gene[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(13): 9043-9048. |
| [12] | Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M, Sato M, Nasu S, Minobe Y. Positional cloning of rice semidwarfing gene, sd-1: Rice "green revolution gene" encodes a mutant enzyme involved in gibberellin synthesis[J]. DNA Research, 2002, 9(1): 11-17. |
| [13] | 李铮友, 师常俊, 李晓艾, 林贤国. 香软米滇屯502的选育[J]. 云南农业大学学报, 1999, 14(1): 27-31. |
| Li Z Y, Shi C J, Li X A, Ling X G. Breeding of Diantun 502: Fragrant and soft rice[J]. Journal of Yunnan Agricultural University, 1999, 14(1): 27-31. (in Chinese with English abstract) | |
| [14] | Biswas S, Tian J Q, Li R, Chen X F, Luo Z J, Chen M J, Zhao X X, Zhang D B, Persson S, Yuan Z, Shi J X. Investigation of CRISPR/Cas9-induced SD1 rice mutants highlights the importance of molecular characterization in plant molecular breeding[J]. Journal of Genetics and Genomics, 2020, 47(5): 273-280. |
| [15] | Hu X, Cui Y, Dong G, Feng A, Wang D, Zhao C, Zhang Y, Hu J, Zeng D, Guo L, Qian Q. Using CRISPR-Cas9 to generate semi-dwarf rice lines in elite landraces[J]. Scientific Reports, 2019, 9(1): 19096. |
| [16] | 胡雪娇, 杨佳, 程灿, 周继华, 牛付安, 王新其, 张美良, 曹黎明, 储黄伟. 利用CRISPR/Cas9系统定向编辑水稻SD1基因[J]. 中国水稻科学, 2018, 32(3): 219-225. |
| Hu X J, Yang J, Cheng C, Zhou J H, Niu F A, Wang X Q, Zhang M L, Cao L M, Chu H W. Targeted editing of rice SD1 gene Using CRISPR/Cas9 system[J]. Chinese Journal of Rice Science, 2018, 32(3): 219-225. (in Chinese with English abstract) | |
| [17] | 黎起秦, 陈育新, 韦绍兴. 赤霉素对稻秆抗瘟性影响的分析[J]. 广西农学院学报, 1990, 9(1): 19-26. |
| Li Q Q, Chen Y X, Wei S X. An analysis on effect of bibberellin on resistance of rice stalk to rice stalk blast[J]. Journal of Guangxi Agricultual College, 1990, 9(1): 19-26. (in Chinese with English abstract) | |
| [18] | Qin X, Liu J H, Zhao W S, Chen X J, Guo Z J, Peng Y L. Gibberellin 20-oxidase gene OsGA20ox3 regulates plant stature and disease development in rice[J]. Molecular Plant-Microbe Interactions, 2013, 26(2): 227-239. |
| [19] | Tanaka N, Matsuoka M, Kitano H, Asano T, Kaku H, Komatsu S. gid1, a gibberellin-insensitive dwarf mutant, shows altered regulation of probenazole-inducible protein (PBZ1) in response to cold stress and pathogen attack[J]. Plant Cell & Environment, 2006, 29(4): 619-631. |
| [20] | Yang D L, Li Q, Deng Y W, Lou Y G, Wang M Y, Zhou G X, Zhang Y Y, He Z H. Altered disease development in the eui mutants and Eui overexpressors indicates that gibberellins negatively regulate rice basal disease resistance[J]. Molecular Plant, 2008, 1(3): 528-537. |
| [21] | Liu H, Ding Y D, Zhou Y Q, Jin W Q, Xie K B, Chen L L. CRISPR-P 2.0: An improved CRISPR-Cas9 tool for genome editing in plants[J]. Molecular Plant, 2017, 10(3): 530-532. |
| [22] | Liu W Z, Xie X R, Ma X L, Li J, Chen J H, Liu Y G. DSDecode: A web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations[J]. Molecular Plant, 2015, 8(9): 1431-1433. |
| [23] | 杨光, 雷海霞, 乔利. 稻瘟病室内产孢及人工接种鉴定[J]. 种业导刊, 2018(12): 23-24. |
| Yang G, Lei H X, Qiao L. Identification of indoor spore production and inoculation of rice blast[J]. Seed Industry Tribune, 2018(12): 23-24. (in Chinese) | |
| [24] | 中华人民共和国农业部. 水稻品种试验稻瘟病抗性鉴定与评价技术规程: NY/T 2646-2014[S]. 北京: 中国标准出版社, 2014: |
| Ministry of Agriculture of the People’s Republic of China. Technical Specification for Identification and Evaluation of Blast Resistance in Rice Variety Region Test: NY/T 2646-2014[S]. Beijing: Standards Press of China, 2014. (in Chinese) | |
| [25] | Arnon D I. Copper enzymes in isolated chloroplasts: Polyphenoloxidase in Beta vulgaris[J]. Plant Physiology, 1949, 24(1): 1-15. |
| [26] | Gao Z Y, Wang Y F, Chen G, Zhang A P, Yang S L, Shang L G, Wang D Y, Ruan B P, Liu C L, Jiang H Z, Dong G J, Zhu L, Hu J, Zhang G H, Zeng D L, Guo L B, Xu G H, Teng S, Harberd N P, Qian Q. The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency[J]. Nature Communication, 2019, 10(1): 5207. |
| [27] | 王新, 韩悦, 冯璇, Nawaz G, 罗亮, 刘芳, 覃宝祥, 刘耀光, 李容柏. 应用 CRISPR-Cas9 基因编辑技术改良传统优质糯稻品种[J]. 分子植物育种, 2019, 17(19): 6332-6342. |
| Wang X, Han Y, Feng X, Nawaz G, Luo L, Liu F, Qin B X, Liu Y G, Li R B. Improvement of a traditional high-quality glutinous rice variety by CRISPR-Cas9 gene editing system[J]. Molecular Plant Breeding, 2019, 17(19): 6332-6342. (in Chinese with English abstract) | |
| [28] | Eshed Y, Lippman Z B. Revolutions in agriculture chart a course for targeted breeding of old and new crops[J]. Science, 2019, 366(6466): eaax0025. |
| [29] | Wang B L, Wei H F, Zhang H, Zhang W H. Enhanced accumulation of gibberellins rendered rice seedlings sensitive to ammonium toxicity[J]. Journal of Experimental Botany, 2020, 71(4): 1514-1526. |
| [30] | Li S, Tian Y H, Wu K, Ye Y F, Yu J P, Zhang J Q, Liu Q, Hu M Y, Li H, Tong Y P, Harberd N P, Fu X D. Modulating plant growth-metabolism coordination for sustainable agriculture[J]. Nature, 2018, 560(7720): 595-600. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||