
中国水稻科学 ›› 2023, Vol. 37 ›› Issue (4): 337-346.DOI: 10.16819/j.1001-7216.2023.221004
• 研究报告 • 下一篇
任志奇1,2, 薛可欣2,3, 董铮2,3, 李小湘2,3, 黎用朝2,3, 郭玉静1,2, 刘文强2,3, 郭梁2,3, 盛新年2,3, 刘之熙2,3, 潘孝武1,2,3( )
)
收稿日期:2022-10-20
修回日期:2022-11-22
出版日期:2023-07-10
发布日期:2023-07-17
通讯作者:
*email: pxw137@163.com
基金资助:
REN Zhiqi1,2, XUE Kexin2,3, DONG Zheng2,3, LI Xiaoxiang2,3, LI Yongzhao2,3, GUO Yujing1,2, LIU Wenqiang2,3, GUO Liang2,3, SHENG Xinnian2,3, LIU Zhixi2,3, PAN Xiaowu1,2,3( )
)
Received:2022-10-20
Revised:2022-11-22
Online:2023-07-10
Published:2023-07-17
Contact:
*email: pxw137@163.com
摘要:
【目的】 适度卷叶可增强叶片的光合效率,提高水稻的产量,筛选和鉴定水稻卷叶突变体有助于解析水稻叶片形成的分子机制。【方法】 利用280 GY钴60 γ射线对籼稻品种玉针香进行诱变处理,筛选出一个外卷叶突变体,暂命名为ocl1(outcurved leaf 1)。考查了突变体的表型特征和农艺性状,并利用ocl1突变体与02428杂交的F2群体定位目标基因,最后通过荧光定量PCR分析了卷叶相关基因的表达情况。【结果】 从分蘖期到成熟期,突变体的叶片表现外卷和披垂;在农艺性状方面,突变体的结实率、千粒重和单株产量显著降低;叶片的显微观察结果表明,突变体的外卷叶表型主要是由于相邻维管束之间的泡状细胞变大引起的;遗传分析表明,ocl1的突变表型受一对隐性核基因控制,将OCL1基因定位在第6染色体上SSR标记RM19575与InDel标记ID02612之间,物理距离约为127 kb;定位区间内的测序结果表明,其中一个基因(LOC_Os06g10600)的内含子-外显子连接处发生单碱基突变,导致异常剪接,进而引起氨基酸序列的改变。该基因编码同源异型域-亮氨酸拉链蛋白,与卷叶相关基因ROC8和URL1等位;荧光定量PCR结果显示,泡状细胞发育相关基因ROC5和LAC17在ocl1突变体中下调表达,而XTH11则在ocl1突变体中上调表达。【结论】 OCL1基因突变通过影响泡状细胞的发育导致叶片外卷,同时引起产量降低。
任志奇, 薛可欣, 董铮, 李小湘, 黎用朝, 郭玉静, 刘文强, 郭梁, 盛新年, 刘之熙, 潘孝武. 水稻外卷叶突变体ocl1的鉴定及基因定位[J]. 中国水稻科学, 2023, 37(4): 337-346.
REN Zhiqi, XUE Kexin, DONG Zheng, LI Xiaoxiang, LI Yongzhao, GUO Yujing, LIU Wenqiang, GUO Liang, SHENG Xinnian, LIU Zhixi, PAN Xiaowu. Identification and Gene Mapping of Outcurved Leaf Mutant ocl1 in Rice[J]. Chinese Journal OF Rice Science, 2023, 37(4): 337-346.
| 引物名称 Primer | 用途 Usage | 正向引物(5'→3') Forward primer (5'→3') | 反向引物(5'→3') Reverse primer (5'→3') |
|---|---|---|---|
| RM217 | 基因定位Gene mapping | ATCGCAGCAATGCCTCGT | GGGTGTGAACAAAGACAC |
| ID02356 | 基因定位Gene mapping | CTCACGTAGGTCTTGAGGAG | AGAAGAGGGCAGGAGGAG |
| RM19570 | 基因定位Gene mapping | CCCAGATATTCTGTGTGATCATGAGG | GAGTGAATGTGAGCCGTCTATTGG |
| RM19575 | 基因定位Gene mapping | TCATCACAAGCTCGTAATCAGG | CCAGAGAATAAGAGGACATGACG |
| ID02612 | 基因定位Gene mapping | GCAGTTAATTATTCCATGCG | TTTGAACTCTCCCATATTCG |
| ID03100 | 基因定位Gene mapping | CCATGGATGACTCTCTCTCT | ACACCTCCACTCCTCCAT |
| RM276 | 基因定位Gene mapping | CTCAACGTTGACACCTCGTG | TCCTCCATCGAGCAGTATCA |
| cDNA-S | 测序Sequencing | AGAGAGGAAGAGGCAGAGGTAG | AGAAGATGCTGTCGTAGGTGTC |
| OsActin | 内参Reference gene | CATTGGTGCTGAGCGTTTCC | AGAAACAAGCAGGAGGACGG |
| RL14 | qRT-PCR | CTCTTTCAGGCATTCCATTGATG | CAACACCTTGTCAGCTTTCAAGC |
| ZHD1 | qRT-PCR | CGAGAACGAATGCTCTCTCAG | CGGACCCCGGTATGGTAG |
| SLL1 | qRT-PCR | GCCTCTGTGATTGCCATCTAAT | CAGGTGTCCAACCATGAGC |
| ROC5 | qRT-PCR | CGCAAGAGGAAGAAGCGATAC | GCTCCAGTTGCGTCTTCATC |
| SRL1 | qRT-PCR | TCTCCTGCCTCCTCTGTGTG | TAGGAGGGGTGGTGTTGAAG |
| NRL1 | qRT-PCR | TCAGTAGTGTAGTGGTGTCGAGTTCA | GCACTCCTTCATGTGAGCTTCA |
| LAC17 | qRT-PCR | CTGCAGATTTGGCACTCGAGAACGTC | CATGCTCTTGGTGTTGCACAG |
| EXPA2 | qRT-PCR | GGGCACTCCTACTTCAACCT | TAGGAGTTGCTCTGCCAGTT |
| EXPA4 | qRT-PCR | GGGCACTCCTACTTCAACCT | CTGGAAGGAGAGGCTCTGG |
| XTH11 | qRT-PCR | ACCTTCTACTTGTCGTCGCA | TGCTGTGGGTTCCAGATGAT |
| CESA2 | qRT-PCR | GGTATCCTTGAGATGAGGTGG | GCCTTTGAGGTGACAGTGAA |
| CESA3 | qRT-PCR | AAGTTCTTCGGTGGGCTCT | TTTCCAGGATGCCAGTAGC |
表1 基因定位、测序及表达分析引物信息
Table 1. Primers for gene mapping, sequencing and expression analysis.
| 引物名称 Primer | 用途 Usage | 正向引物(5'→3') Forward primer (5'→3') | 反向引物(5'→3') Reverse primer (5'→3') |
|---|---|---|---|
| RM217 | 基因定位Gene mapping | ATCGCAGCAATGCCTCGT | GGGTGTGAACAAAGACAC |
| ID02356 | 基因定位Gene mapping | CTCACGTAGGTCTTGAGGAG | AGAAGAGGGCAGGAGGAG |
| RM19570 | 基因定位Gene mapping | CCCAGATATTCTGTGTGATCATGAGG | GAGTGAATGTGAGCCGTCTATTGG |
| RM19575 | 基因定位Gene mapping | TCATCACAAGCTCGTAATCAGG | CCAGAGAATAAGAGGACATGACG |
| ID02612 | 基因定位Gene mapping | GCAGTTAATTATTCCATGCG | TTTGAACTCTCCCATATTCG |
| ID03100 | 基因定位Gene mapping | CCATGGATGACTCTCTCTCT | ACACCTCCACTCCTCCAT |
| RM276 | 基因定位Gene mapping | CTCAACGTTGACACCTCGTG | TCCTCCATCGAGCAGTATCA |
| cDNA-S | 测序Sequencing | AGAGAGGAAGAGGCAGAGGTAG | AGAAGATGCTGTCGTAGGTGTC |
| OsActin | 内参Reference gene | CATTGGTGCTGAGCGTTTCC | AGAAACAAGCAGGAGGACGG |
| RL14 | qRT-PCR | CTCTTTCAGGCATTCCATTGATG | CAACACCTTGTCAGCTTTCAAGC |
| ZHD1 | qRT-PCR | CGAGAACGAATGCTCTCTCAG | CGGACCCCGGTATGGTAG |
| SLL1 | qRT-PCR | GCCTCTGTGATTGCCATCTAAT | CAGGTGTCCAACCATGAGC |
| ROC5 | qRT-PCR | CGCAAGAGGAAGAAGCGATAC | GCTCCAGTTGCGTCTTCATC |
| SRL1 | qRT-PCR | TCTCCTGCCTCCTCTGTGTG | TAGGAGGGGTGGTGTTGAAG |
| NRL1 | qRT-PCR | TCAGTAGTGTAGTGGTGTCGAGTTCA | GCACTCCTTCATGTGAGCTTCA |
| LAC17 | qRT-PCR | CTGCAGATTTGGCACTCGAGAACGTC | CATGCTCTTGGTGTTGCACAG |
| EXPA2 | qRT-PCR | GGGCACTCCTACTTCAACCT | TAGGAGTTGCTCTGCCAGTT |
| EXPA4 | qRT-PCR | GGGCACTCCTACTTCAACCT | CTGGAAGGAGAGGCTCTGG |
| XTH11 | qRT-PCR | ACCTTCTACTTGTCGTCGCA | TGCTGTGGGTTCCAGATGAT |
| CESA2 | qRT-PCR | GGTATCCTTGAGATGAGGTGG | GCCTTTGAGGTGACAGTGAA |
| CESA3 | qRT-PCR | AAGTTCTTCGGTGGGCTCT | TTTCCAGGATGCCAGTAGC |
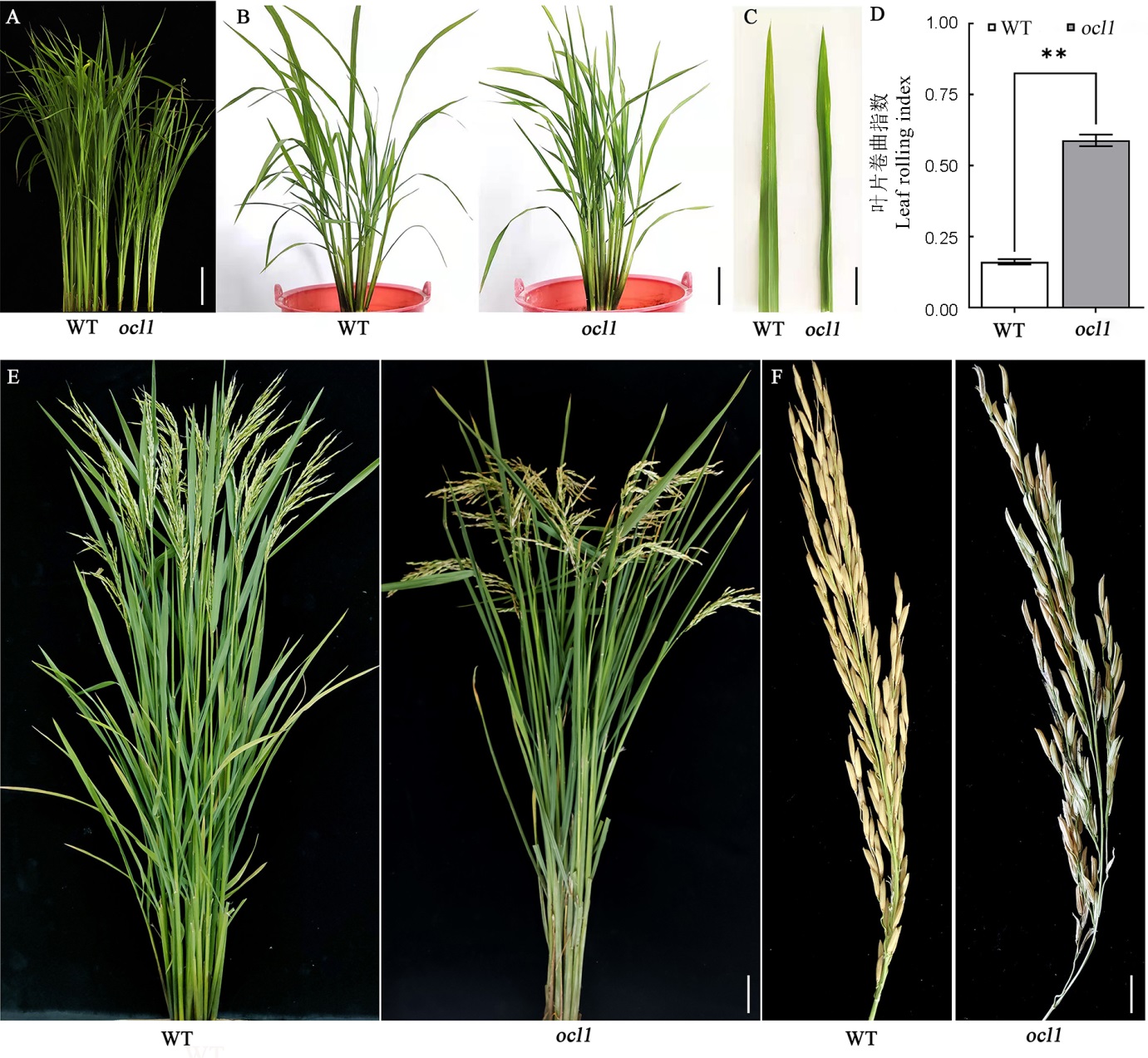
图1 野生型与ocl1突变体的表型 A—野生型(WT)与突变体在苗期的植株形态,标尺为5 cm;B—野生型和突变体在分蘖期的植株形态,标尺为15 cm;C—野生型与突变体在分蘖期的叶片形态, 标尺为1 cm;D—野生型与突变体在分蘖期的叶片卷曲指数;E—野生型与突变体抽穗期的植株形态,标尺为10 cm;F—野生型和突变体的结实情况,标尺为2 cm。
Fig. 1. Plant architecture of the wild type and ocl1. A, Plant architecture of WT and ocl1 at the seedling stage, bar=5 cm; B, Plant architecture of WT and ocl1 at the tillering stage, bar=15 cm; C, Leaf morphology of WT and ocl1 at the tillering stage, bar=1 cm; D, Leaf rolling index of WT and ocl1 at the tillering stage; E, Plant architecture of WT and ocl1 at the heading stage, bar=10 cm; F, Seed setting of WT and ocl1, bar=2 cm.
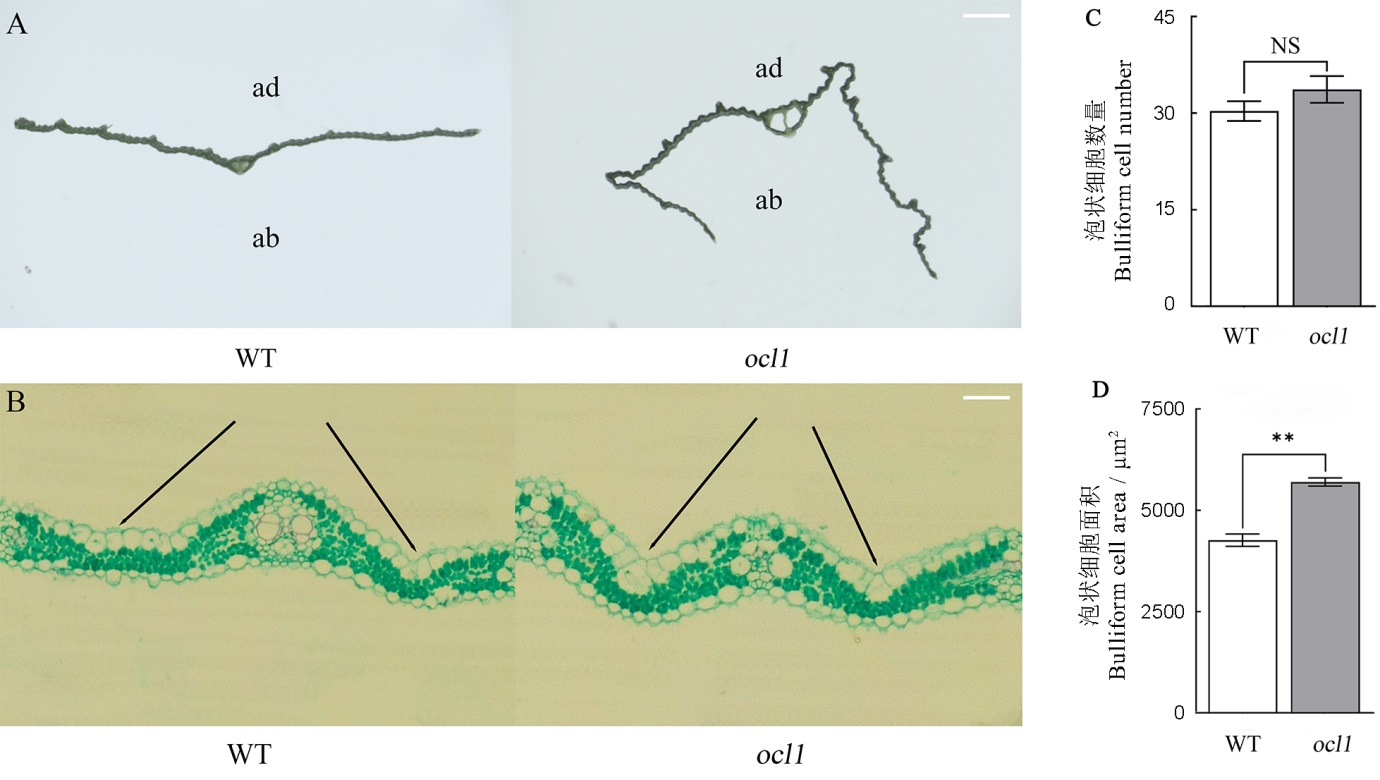
图2 野生型与ocl1突变体叶片的横截面及切片观察 A―野生型与ocl1突变体的横截面观察,标尺为1 mm;B―野生型与ocl1突变体的切片观察,黑色线条指向野生型与ocl1突变体的泡状细胞,标尺为100 μm;C―野生型与突变体的泡状细胞数量; D―野生型与突变体的泡状细胞面积;NS表示突变体与野生型之间无显著性差异。**表示突变体与野生型之间的差异达0.01显著水平。
Fig. 2. Cross-sectional and leaf slice observation of WT and ocl1. A, Cross-sectional observation of WT and ocl1, bar=1 mm; B, Leaf slice observation of WT and ocl1, black lines point to bulliform cells of WT and ocl1, bar=100 μm; C, Number of bulliform cells of WT and ocl1; D, Area of bulliform cells of WT and ocl1; NS means there is no significant difference between the mutant and WT; ** Significant difference between the mutant and WT at 0.01 level. ad, Adaxial side; ab, Abaxial side.
| 材料 Material | 单株穗数 | 每穗总粒数 | 结实率 | 千粒重 | 单株产量 | |
|---|---|---|---|---|---|---|
| No. of panicles per plant | No. of spikelets per panicle | Seed-setting rate / % | 1000-grain weight / g | Yield per plant / g | ||
| WT | 11.9±0.4 | 94.2±1.3 | 74.4±0.8 | 29.6±0.1 | 24.7±1.1 | |
| ocl1 | 11.3±0.1 | 92.4±4.0 | 45.7±2.0** | 24.8±0.1** | 11.8±0.7** | |
表2 野生型与突变体ocl1的主要农艺性状比较
Table 2. Agronomic traits of the WT and ocl1.
| 材料 Material | 单株穗数 | 每穗总粒数 | 结实率 | 千粒重 | 单株产量 | |
|---|---|---|---|---|---|---|
| No. of panicles per plant | No. of spikelets per panicle | Seed-setting rate / % | 1000-grain weight / g | Yield per plant / g | ||
| WT | 11.9±0.4 | 94.2±1.3 | 74.4±0.8 | 29.6±0.1 | 24.7±1.1 | |
| ocl1 | 11.3±0.1 | 92.4±4.0 | 45.7±2.0** | 24.8±0.1** | 11.8±0.7** | |

图3 野生型与ocl1突变体的籽粒形态分析 A―野生型(WT)与突变体的粒长,标尺为1 cm;B―野生型和突变体的粒宽,标尺为0.5 cm;C―野生型与突变体的糙米饱满度,标尺为1 cm;D―野生型与突变体的粒长、粒宽和粒厚统计学分析。**表示突变体与野生型之间的差异达0.01显著水平。
Fig. 3. Analysis of grain morphology between WT and ocl1. A, Grain length of WT and ocl1, bar=1 cm; B, Grain width of WT and ocl1, bar=5 mm; C, Grain plumpness of WT and ocl1, bar=1 cm; D, Statistical analysis of grain length, grain width and grain thickness of WT and ocl1; **Significant difference between the mutant and WT at 0.01 level.

图4 OCL1基因的图位克隆 A—水稻OCL1基因的定位,黑色方框代表外显子,白色方框代表UTR,黑色线条代表内含子,黑色箭头表示突变位置;B—水稻OCL1基因的突变位点;C—WT与ocl1的cDNA扩增产物;D—WT与ocl1的cDNA序列比对结果;E—氨基酸序列比对结果。
Fig. 4. Map-based cloning of OCL1. A, Mapping of OCL1 gene. Black boxes, white boxes and black lines represent exons, UTR and introns, respectively. Black arrow indicates the mutation site; B, Sequence comparison of WT and ocl1 at the mutation site; C, Electrophoresis detection of cDNA amplification products; D, cDNA sequence alignment of the OCL1 gene between WT and ocl1; E, Alignment of the amino acid sequence between WT and ocl1.
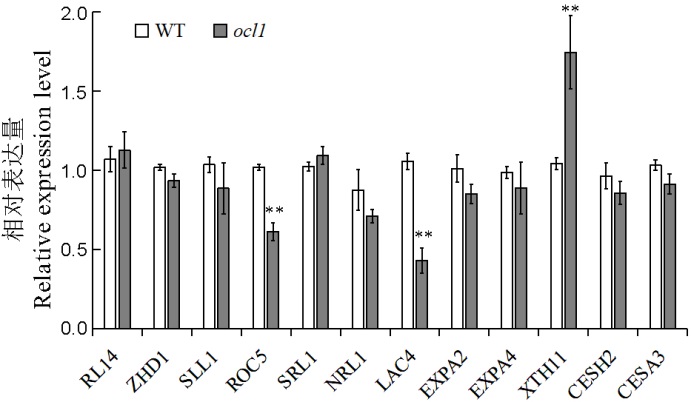
图5 水稻卷叶相关基因的相对表达量 **表示突变体与野生型之间的差异达0.01显著水平。
Fig. 5. Relative expression level of genes associated with leaf rolling. **Significant difference between the mutant and WT at 0.01 level.
| [1] | 刘永巍, 田红刚, 李春光, 孟昭河, 程芳艳, 孙翊轩, 刘忠良. 水稻超高产育种的研究[J]. 植物学研究, 2014, 3(4): 172-177. |
| Liu Y W, Tian H G, Li C G, Meng Z H, Cheng F Y, Sun Y X, Liu Z L. The study on super high yield breeding of rice[J]. Botanical Research, 2014, 3(4): 172-177. (in Chinese with English abstract) | |
| [2] | Xu Y, Ma K, Zhao Y, Wang X, Zhou K, Yu G, Li C, Li P, Yang Z, Xu C. Genomic selection: A breakthrough technology in rice breeding[J]. The Crop Journal, 2021, 9(3): 669-677. |
| [3] | 郭韬, 余泓, 邱杰, 李家洋, 韩斌, 林鸿宣. 中国水稻遗传学研究进展与分子设计育种[J]. 中国科学: 生命科学, 2019, 49(10): 1185-1212. |
| Guo T, Yu H, Qiu J, Li J X, Han B, Lin H X. Advances in rice genetics and breeding by molecular design in China[J]. Science China: Life Sciences, 2019, 49(10): 1185-1212. (in Chinese with English abstract) | |
| [4] | 梁程, 向珣朝, 张欧玲, 游慧, 许亮, 陈永军. 两份新株型水稻品系的农艺性状与遗传特性分析[J]. 中国水稻科学, 2022, 36(2): 171-180. |
| Liang C, Xiang X C, Zhang O L, You H, Xu L, Chen Y J. Analyses on agronomic traits and genetic characteristics of two new plant-architecture lines in rice[J]. Chinese Journal of Rice Science, 2022, 36(2): 171-180. (in Chinese with English abstract) | |
| [5] | 刘坚, 陶红剑, 施思, 叶卫军, 钱前, 郭龙彪. 水稻穗型的遗传和育种改良[J]. 中国水稻科学, 2012, 26(2): 227-234. |
| Liu J, Tao H J, Shi S, Ye W J, Qian Q, Guo L B. Genetics and breeding improvement for panicle type in rice[J]. Chinese Journal of Rice Science, 2012, 26(2): 227-234. (in Chinese with English abstract) | |
| [6] | Luo Y, Zhao F, Sang X, Ling Y, Yang Z, He G. Genetic analysis and gene mapping of a novel rolled-leaf mutant rl12(t) in rice[J]. Acta Agronomica Sinica, 2009, 35(11): 1967-1972. |
| [7] | Shi Z, Wang J, Wan X, Shen G, Wang X, Zhang J. Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit[J]. Planta, 2007, 226(1): 99-108. |
| [8] | Fujino K, Matsuda Y, Ozawa K, Nishimura T, Koshiba T, Marco W, Sekiguchi H. Narrow leaf7 controls leaf shape mediated by auxin in rice[J]. Molecular Genetics & Genomics, 2008, 279(5): 499-507. |
| [9] | Zhang G, Xu Q, Zhu X, Qian Q, Xue H. SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development[J]. The Plant Cell, 2009, 21(3): 719-735. |
| [10] | Fang L, Zhao F, Cong Y, Sang X, Du Q, Wang D, Li Y, Ling Y, Yang Z, He G. Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves[J]. Plant Biotechnology Journal, 2012, 10(5): 524-532. |
| [11] | Itoh J I, Nonomura K I, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y. Rice plant development: from zygote to spikelet[J]. Plant Cell Physiology, 2005, 46(1): 23-47. |
| [12] | Xiang J, Zhang G, Qian Q, Xue H. SEMI-ROLLED LEAF1 encodes a putative glycosylphosphatidylinositol- anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells[J]. Plant Physiology, 2012, 159(4): 1488-1500. |
| [13] | Sun J, Cui X, Teng S, Kunnong Z, Wang Y, Chen Z, Sun X, Wu J, Ai P, Quick W P, Lu T, Zhang Z. HD-ZIP IV gene Roc8 regulates the size of bulliform cells and lignin content in rice[J]. Plant Biotechnology Journal, 2020, 18(12): 2559-2572. |
| [14] | Zou L, Sun X, Zhang Z, Liu P, Wu J, Tian C, Qiu J, Lu T. Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice[J]. Plant Physiology, 2011, 156(3): 1589-1602. |
| [15] | Li L, Shi Z, Li L, Shen G, Wang X, An L S, Zhang J. Overexpression of ACL1 (abaxially curled leaf 1) increased bulliform cells and induced abaxial curling of leaf blades in rice[J]. Molecular Plant, 2010, 3(5): 807-817. |
| [16] | Xu Y, Kong W, Wang F, Wang J, Tao Y, Li W, Chen Z, Fan F, Jiang Y, Zhu Q, Yang J. Heterodimer formed by ROC8 and ROC5 modulates leaf rolling in rice[J]. Plant Physiology, 2021, 19(12): 2662-2672. |
| [17] | Fang J, Guo T, Xie Z, Chun Y, Zhao J, Peng L, Zafar S A, Yuan S, Xiao L, Li X. The URL1-ROC5-TPL2 transcriptional repressor complex represses the ACL1 gene to modulate leaf rolling in rice[J]. Plant Physiology, 2021, 185(4): 1722-1744. |
| [18] | Hibara K I, Obara M, Hayashida E, Abe M, Ishimaru T, Satoh H, Itoh J, Nagato Y. The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice[J]. Developmental Biology, 2009, 334(2): 345-354. |
| [19] | Chen Q, Xie Q, Gao J, Wang W Y, Sun B, Liu B, Zhu H, Peng H, Zhao H, Liu C, Wang J, Zhang J, Zhang G, Zhang Z. Characterization of rolled and erect leaf 1 in regulating leave morphology in rice[J]. Journal of Experimental Botany, 2015, 66(19): 6047-6058. |
| [20] | Xu Y, Wang Y, Long Q, Huang J, Wang Y, Zhou K, Zheng M, Sun J, Chen S H, Jiang L, Wang C M, Wan J. Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice[J]. Planta, 2014, 239(4): 803-816. |
| [21] | 谢园华, 李凤菲, 马晓慧, 谭佳, 夏赛赛, 桑贤春, 杨正林, 凌英华. 水稻半外卷叶突变体sol1 的表型分析与基因定位[J]. 作物学报, 2020, 46(2): 204-213. |
| Xie Y H, Li F F, Ma X H, Tan J, Xia S S, Sang X C, Yang Z L, Ling Y H. Phenotype characterization and gene mapping of the semi-outcurved leaf mutant sol1 in rice (Oryza sativa L.)[J]. Acta Agronomica Sinica, 2020, 46(2): 204-213. (in Chinese with English abstract) | |
| [22] | 刘强, 张贵友, 陈受宜. 植物转录因子的结构与调控作用[J]. 科学通报, 2000, 45(14): 1465-1474. |
| Liu Q, Zhang G Y, Chen S Y. Structure and regulation of plant transcription factors[J]. Chinese Science Bulletin, 2000, 45(14): 1465-1474. (in Chinese with English abstract) | |
| [23] | 吴方喜, 罗曦, 蒋家焕, 连玲, 魏毅东, 何炜, 陈丽萍, 蔡秋华, 谢华安, 张建福. 水稻卷叶突变体基因 shallot like1-Fuhui673 鉴定, 克隆与序列分析[J]. 科学通报, 2018, 63(23): 2369-2377. |
| Wu F X, Luo X, Jiang J H, Lian L, Wei Y D, He W, Chen L P, Cai Q H, Xie H A, Zhang J F. Identification, cloning and sequence analysis shallot like1-Fuhui673 a rolled leaf mutant in rice[J]. Chinese Science Bulletin, 2018, 63(23): 2369-2377. (in Chinese with English abstract) | |
| [24] | Eshed Y, Izhaki A, Baum S F, Floyd S K, Bowman J L. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities[J]. Development, 2004, 131(12): 2997-3006. |
| [25] | Wang B, Smith S M, Li J. Genetic regulation of shoot architecture[J]. Annual Review of Plant Biology, 2018, 69: 437-468. |
| [26] | 邓秋雨, 肖应辉. 水稻卷叶类型及调控机制研究进展[J]. 作物研究, 2021, 35(4): 376-384. |
| Deng Q Y, Xiao Y H. Research progress on types and regulation mechanism of rice rolled leaf[J]. Crop Research, 2021, 35(4): 376-384. (in Chinese with English abstract) | |
| [27] | 张小惠, 秦亚芝, 张迎信, 占小登, 张振华, 沈希宏, 程式华, 曹立勇, 吴先军. 水稻窄卷叶突变体Nrl3(t)的基因定位[J]. 中国水稻科学, 2015, 29(6): 595-600. |
| Zhang X H, Qin Y Z, Zhang Y X, Zhan X D, Zhang Z H, Shen X H, Cheng S H, Cao L Y, Wu X J. Gene mapping of a narrow and rolled leaf mutant Nrl3(t) in rice[J]. Chinese Journal of Rice Science, 2015, 29(6): 595-600. (in Chinese with English abstract) | |
| [28] | Luan W, Liu Y, Zhang F, Song Y, Wang Z, Peng Y, Sun Z. OsCD1 encodes a putative member of the cellulose synthase-like D sub-family and is essential for rice plant architecture and growth[J]. Plant Biotechnology Journal, 2010, 9(4): 513-524. |
| [29] | 赵芳明, 魏霞, 马玲, 桑贤春, 王楠, 张长伟, 凌英华, 何光华. 水稻生育后期卷叶突变体lrl1的鉴定及基因定位和候选基因预测[J]. 科学通报, 2015, 60(32): 3133-3143. |
| Zhao F M, Wei X, Ma L, Sang X C, Wang N, Zhang C W, Ling Y H, He G H. Identification, gene mapping and candidate gene prediction of a late-stage rolled leaf mutant lrl1 in rice (Oryza sativa L.)[J]. Chinese Science Bulletin, 2015, 60(32): 3133-3143. (in Chinese with English abstract) | |
| [30] | 刘晨, 孔维一, 尤世民, 钟秀娟, 江玲, 赵志刚, 万建民. 一个水稻卷叶基因的遗传分析和精细定位[J]. 中国农业科学, 2015, 48(13): 2487-2496. |
| Liu C, Kong W Y, You L M, Zhong X J, Jiang L, Zhao Z G, Wan J M. Genetic analysis and fine mapping of a novel rolled leaf gene in rice[J]. Scientia Agricultura Sinica, 2015, 48(13): 2487-2496. (in Chinese with English abstract) | |
| [31] | Wang D, Liu H, Li K, Li S, Tao Y. Genetic analysis and gene mapping of a narrow leaf mutant in rice (Oryza sativa L)[J]. Chinese Science Bulletin, 2009, 54: 752-758. |
| [32] | Li C, Zou X, Zhang C, Shao Q, Liu J, Liu B, Li H, Zhao T. OsLBD3-7 overexpression induced adaxially rolled leaves in rice[J]. PLoS One, 2016, 11(6): e0156413. |
| [33] | Li W, Zhang M, Gan P, Qian L, Yang S, Miao, Wang G, Zhang M, Liu W, Li H, Shi C, Chen K. CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice[J]. The Plant Journal, 2017, 92(5): 904-923. |
| [34] | Jan A, Yang G, Nakamura H, Ichikawa H, Kitano H, Matsuoka M, Matsumoto H, Komatsu S. Characterization of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice[J]. Plant Physiology, 2004, 136(3): 3670-3681. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||