
中国水稻科学 ›› 2023, Vol. 37 ›› Issue (1): 37-44.DOI: 10.16819/j.1001-7216.2023.220203
袁洋1,2, 敖和军1, 周仲华1, 应杰政2, 张健2, 倪深2( )
)
收稿日期:2022-02-02
修回日期:2022-04-01
出版日期:2023-01-10
发布日期:2023-01-10
通讯作者:
倪深
基金资助:
YUAN Yang1,2, AO Hejun1, ZHOU Zhonghua1, YING Jiezheng2, ZHANG Jian2, NI Shen2( )
)
Received:2022-02-02
Revised:2022-04-01
Online:2023-01-10
Published:2023-01-10
Contact:
NI Shen
摘要: 目的 通过研究水稻渐渗系群体构建时产生的偏分离现象,可以发现更多与偏分离相关的基因,有助于解析偏分离在水稻中的主要作用及其产生机制。方法 应用籼粳组合华占/Koliya构建的一套渐渗系群体,以其后代基因型的分离比作为表型对偏分离位点进行定位。结果 在第8染色体上发现一个染色体区段在其后代中的基因型分离比偏离孟德尔分离定律。进一步分析表明该偏分离现象受第8染色体上两个位点控制,分别命名为SegD8A和SegD8B,其中SegD8A为偏分离位点,但其偏分离属性受SegD8B调控。当SegD8A和SegD8B位点都为杂合型时,子代中SegD8AHH和SegD8AKK的比例接近1∶4;当SegD8B为纯合华占基因型时,子代中SegD8AHH和SegD8AKK的比例接近1∶1;当SegD8B为纯合Koliya基因型时,子代中SegD8AHH几乎不出现。我们将SegD8A基因定位在一个大小为46.5 kb的区间内,SegD8B基因定位在一个大小为1160.2 kb的区间内。结论 本研究为进一步克隆这两个基因,并阐明该偏分离现象奠定了基础。
袁洋, 敖和军, 周仲华, 应杰政, 张健, 倪深. 水稻杂种偏分离位点SegD8的精细定位[J]. 中国水稻科学, 2023, 37(1): 37-44.
YUAN Yang, AO Hejun, ZHOU Zhonghua, YING Jiezheng, ZHANG Jian, NI Shen. Fine-mapping of SegD8 Loci for Rice Hybrid Segregation Distortion[J]. Chinese Journal OF Rice Science, 2023, 37(1): 37-44.
| 标记 Marker | 基因型 Genotype | χ2(1:2:1) | ||
|---|---|---|---|---|
| HZ/HZ | HZ/BLY | BLY/BLY | ||
| CHR8-24 | 6 | 35 | 29 | 15.11** |
| CHR8-22 | 7 | 40 | 23 | 8.74* |
| CHR8-5 | 8 | 41 | 21 | 6.89* |
| CHR8-25 | 9 | 36 | 25 | 7.37* |
表1 分子标记在第8染色体上的偏分离
Table 1. Segregation distortion of molecular markers on chromosome 8.
| 标记 Marker | 基因型 Genotype | χ2(1:2:1) | ||
|---|---|---|---|---|
| HZ/HZ | HZ/BLY | BLY/BLY | ||
| CHR8-24 | 6 | 35 | 29 | 15.11** |
| CHR8-22 | 7 | 40 | 23 | 8.74* |
| CHR8-5 | 8 | 41 | 21 | 6.89* |
| CHR8-25 | 9 | 36 | 25 | 7.37* |
| 正常花粉率Normal pollen rate/% | |
| 92.56 | |
| 92.35ns | |
| 92.11ns | |
| 92.41ns | |
| 7.88 | |
表2 自交群体偏分离株系正常花粉率与结实率
Table 2 Normal pollen rates and setting rates of segregation distortion in inbred populations.
| 正常花粉率Normal pollen rate/% | |
| 92.56 | |
| 92.35ns | |
| 92.11ns | |
| 92.41ns | |
| 7.88 | |
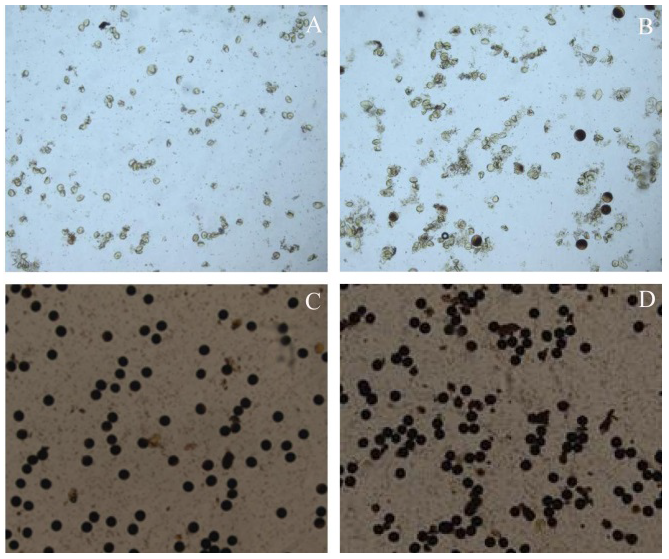
图2 BC3F2代群体花粉染色图 A―正常子代低温不育; B―偏分离子代低温不育; C―子代正常分离; D―子代偏分离。
Fig. 2. Pictures of pollen stained of BC3F2 population. A, Sterility of normally-segregated offspring at low temperature; B, Sterility of offspring with segregation distortion at low temperature; C, Normal segregation of offspring; D, Segregation distortion of offspring.
| 单株 Plant | 标记 Marker | 基因型 Genotype | 分离比Segregation ratio (HZ/HZ∶HZ/BLY∶BLY/BLY) | ||
|---|---|---|---|---|---|
| HZ/HZ | HZ/BLY | BLY/BLY | |||
| NFX9-56-4 | CHR8-45 | 5 | 26 | 16 | 1∶5∶4 |
| NFX9-80-1 | CHR8-45 | 4 | 24 | 20 | |
| NFX10-128-5 | CHR8-45 | 5 | 25 | 18 | |
| NFX9-34-1 | CHR8-45 | 1 | 24 | 23 | 0∶1∶1 |
| NFX9-37-6 | CHR8-45 | 0 | 25 | 23 | |
| NFX10-113-1 | CHR8-45 | 1 | 20 | 27 | |
| NFX9-26-4 | CHR8-45 | 12 | 24 | 12 | 1∶2∶1 |
| NFX10-130-6 | CHR8-45 | 13 | 24 | 10 | |
| NFX6-13-6 | CHR8-45 | 21 | 52 | 21 | |
表3 群体后代偏分离比率的类型
Table 3. Types of partial segregation ratio of population offspring.
| 单株 Plant | 标记 Marker | 基因型 Genotype | 分离比Segregation ratio (HZ/HZ∶HZ/BLY∶BLY/BLY) | ||
|---|---|---|---|---|---|
| HZ/HZ | HZ/BLY | BLY/BLY | |||
| NFX9-56-4 | CHR8-45 | 5 | 26 | 16 | 1∶5∶4 |
| NFX9-80-1 | CHR8-45 | 4 | 24 | 20 | |
| NFX10-128-5 | CHR8-45 | 5 | 25 | 18 | |
| NFX9-34-1 | CHR8-45 | 1 | 24 | 23 | 0∶1∶1 |
| NFX9-37-6 | CHR8-45 | 0 | 25 | 23 | |
| NFX10-113-1 | CHR8-45 | 1 | 20 | 27 | |
| NFX9-26-4 | CHR8-45 | 12 | 24 | 12 | 1∶2∶1 |
| NFX10-130-6 | CHR8-45 | 13 | 24 | 10 | |
| NFX6-13-6 | CHR8-45 | 21 | 52 | 21 | |
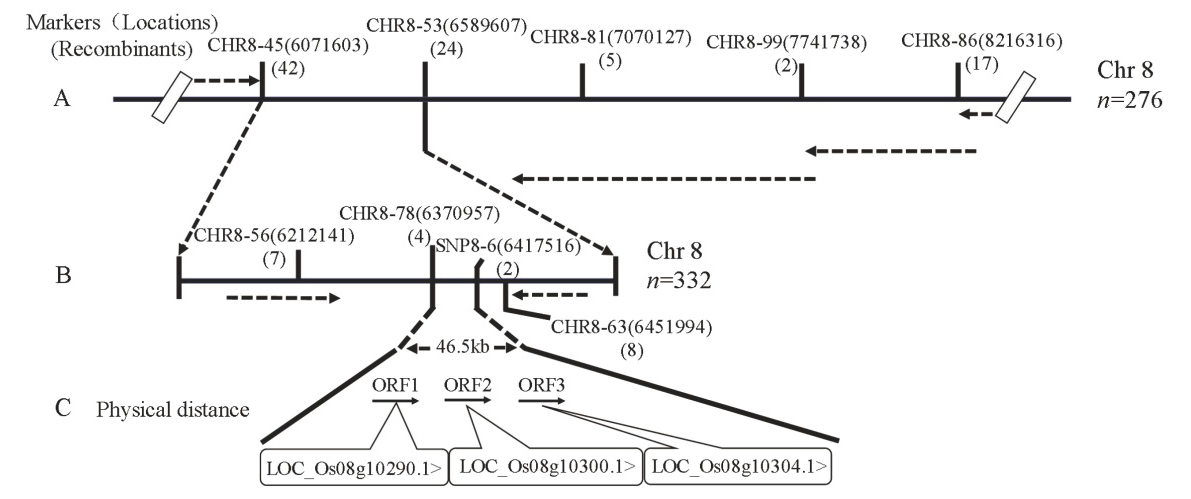
图3 SegD8A在水稻第8染色体上的精细定位 A―SegD8A初定位在第8染色体CHR8-45和CHR8-53之间; B―SegD8A精细定位在CHR8-78和SNP8-6之间46.5 kb区间内; C―精细定位区间内包含的开放阅读框(open reading frame, ORFs)和预测基因。
Fig. 3. Fine mapping of SegD8A on chromosome 8 of rice. A, Primary mapping of SegD8A between markers CHR8-45 and CHR8-53 on chromosome 8; B, SegD8A was fine mapped to the interval between markers CHR8-78 and SNP8-6, a 46.5 kb region; C, Open reading frames (ORFs) and predictive genes within the fine-mapped region.
| 标记 Marker | 类型 Type | 正向引物(5'-3') Forward primer(5'-3') | 反向引物(5'-3') Reverse primer(5'-3') |
| CHR8-45 | InDel | TTAGCATGGAGTTTATTTTTAGTGAT | ACCATGACCACCTTCGCA |
| CHR8-56 | InDel | GACCCTCCTATTTGCCCTTGT | GTTGTTTGTCTTTTGTGGACTGTG |
| CHR8-78 | InDel | CCAAAGAAATTGCCAATCCTAC | CTATGTTGTGTTTCCTGAAATCACA |
| SNP8-6 | SNP | CCAAGCGTTTGCTCACTGC | TACTCTATCCCTTCTTTCTTCGCTT |
| CHR8-63 | InDel | GAGTAACTTTTTTTATGTCGCCAA | ACCTGGTTTCGCAAGTTCG |
| CHR8-53 | InDel | CGAGCAGACCAATCTAAGGGAT | AACAGAAGACCCCAGAGAATCAA |
| CHR8-71 | InDel | GGAAAATAAAAACAAAAGACGGAAC | CATCAGAGGAGGAGTACTTCAGTGA |
| CHR8-80 | InDel | GCTAAACTTTCCTGTAACATCCTGA | GAAAATGGTTTCAATTTCTACCCA |
| CHR8-29 | InDel | ATACGCAGAGCCATATATAAAAGAT | CTACACGCACAAACTATTTCAGTAC |
| CHR8-86 | InDel | AGCAGGCACGGGCGATTA | TGGAGCATTTGGCAGGGTTA |
表4 本研究使用的标记
Table 4. DNA markers used in this study.
| 标记 Marker | 类型 Type | 正向引物(5'-3') Forward primer(5'-3') | 反向引物(5'-3') Reverse primer(5'-3') |
| CHR8-45 | InDel | TTAGCATGGAGTTTATTTTTAGTGAT | ACCATGACCACCTTCGCA |
| CHR8-56 | InDel | GACCCTCCTATTTGCCCTTGT | GTTGTTTGTCTTTTGTGGACTGTG |
| CHR8-78 | InDel | CCAAAGAAATTGCCAATCCTAC | CTATGTTGTGTTTCCTGAAATCACA |
| SNP8-6 | SNP | CCAAGCGTTTGCTCACTGC | TACTCTATCCCTTCTTTCTTCGCTT |
| CHR8-63 | InDel | GAGTAACTTTTTTTATGTCGCCAA | ACCTGGTTTCGCAAGTTCG |
| CHR8-53 | InDel | CGAGCAGACCAATCTAAGGGAT | AACAGAAGACCCCAGAGAATCAA |
| CHR8-71 | InDel | GGAAAATAAAAACAAAAGACGGAAC | CATCAGAGGAGGAGTACTTCAGTGA |
| CHR8-80 | InDel | GCTAAACTTTCCTGTAACATCCTGA | GAAAATGGTTTCAATTTCTACCCA |
| CHR8-29 | InDel | ATACGCAGAGCCATATATAAAAGAT | CTACACGCACAAACTATTTCAGTAC |
| CHR8-86 | InDel | AGCAGGCACGGGCGATTA | TGGAGCATTTGGCAGGGTTA |
| [1] |
Sano Y. The genic nature of gamete eliminator in rice[J]. Genetics, 1990, 125(1): 183-191.
PMID |
| [2] |
Lu H, Romero-Severson J, Bernardo R. Chromosomal regions associated with segregation distortion in maize[J]. Theoretical and Applied Genetics, 2002, 105(4): 622-628.
PMID |
| [3] | Ellstrand N C, Schierenbeck K A. Hybridization as a stimulus for the evolution of invasiveness in plants?[J]. Proceedings of the National Academy of Sciences, 2000, 97(13): 7043-7050. |
| [4] |
Taylor D R, Ingvarsson P K. Common features of segregation distortion in plants and animals[J]. Genetica, 2003, 117(1): 27-35.
PMID |
| [5] |
Xia F, Ouyang Y D. Recurrent breakdown and rebalance of segregation distortion in the genomes: battle for the transmission advantage[J]. aBIOTECH, 2020, 1: 246-254
PMID |
| [6] |
Xu Y, Zhu L, Xiao J, Huang N, McCouch S R. Chromosomal regions associated with segregation distortion of molecular markers in F2, backcross, doubled haploid, and recombinant inbred populations in rice (Oryza sativa L.)[J]. Molecular and General Genetics, 1997, 253(5): 535-545.
PMID |
| [7] |
Reinisch A J, Dong J M, Brubaker C L, Stelly D M, Wendel J F, Paterson A H. A detailed RFLP map of cotton, Gossypium hirsutum × Gossypium barbadense: Chromosome organization and evolution in a disomic polyploid genome[J]. Genetics, 1994, 138(3): 829-847.
PMID |
| [8] | Matsushita S, Iseki T, Fukuta Y, Araki E, Kobayashi S, Osaki M, Yamagishi M. Characterization of segregation distortion on chromosome 3 induced in wide hybridization between indica and japonica type rice varieties[J]. Euphytica, 2003, 134(1): 27-32. |
| [9] | Sibov S T, Lopes De Souza Jr C, Garcia A F. Molecular mapping in tropical maize (Zea mays L.) using microsatellite markers: Map construction and localization of loci showing distorted segregation[J]. Hereditas, 2003, 139(2): 96-106. |
| [10] | Song X L, Sun X Z, Zhang T Z. Segregation distortion and its effect on genetic mapping in plants[J]. Chinese Journal of Agricultural Biotechnology, 2006, 3(3): 163-169. |
| [11] |
Wang G, He Q, Xu Z K, Song R T. High segregation distortion in maize B73×Teosinte crosses[J]. Genetics and Molecular Research, 2012, 11(1): 693-706.
PMID |
| [12] |
Giesbers A K J, den Boer E,Ulen J J W E H,van Kaauwen M P W,Visser R G F,Niks R E,Jeuken M J W. Patterns of transmission ratio distortion in interspecific lettuce hybrids reveal a sex-independent gametophytic barrier[J]. Genetics, 2019, 211(1): 263-276.
PMID |
| [13] |
Luo D P, Xu H, Liu Z L, Guo J X, Li H Y, Chen L T, Fang C, Zhang Q Y, Bai M, Yao N, Wu H, Wu H, Ji C H, Zheng H Q, Chen Y L, Ye S, Li X Y, Zhao X C, Li R Q, Liu Y G. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice[J]. Nature Genetics, 2013, 45(5): 573-577.
PMID |
| [14] | Wang S H, Tan Y L, Tan X L, Zhang Z L, Wen J C, Kou S Y. Segregation distortion detected in six rice F2 populations generated from reciprocal hybrids at three altitudes[J]. Genetics Research, 2009, 91(5): 345-353. |
| [15] | 姚焱. 花粉愈伤组织群体偏态分离的 SSR 标记鉴定[J]. 广州大学学报, 2003, 2(6): 512-514. |
| Yao Y. Detection of distorted segregation in pollen callus population using SSR marker[J]. Journal of Guangzhou University: Natural Science Edition, 2003, 2(6): 512-514. (in Chinese with English abstract) | |
| [16] | Flachs P, Mihola O, Šimeček P, Gregorova S. Hst1)gene in mouse hybrid sterility[J]. PLoS Genetics, 2012, 8(11): e1003044. |
| [17] |
Pimpinelli S, Dimitri P. Cytogenetic analysis of segregation distortion in Drosophila melanogaster: The cytological organization of the Responder (Rsp) locus[J]. Genetics, 1989, 121(4): 765-772.
PMID |
| [18] |
Yang J Y, Zhao X B, Cheng K, Du H Y, Ouyang Y D, Chen J J, Qiu S Q, Huang J Y, Jiang Y H, Jiang L W, Ding J H, Wang J, Xu C Q, Li X H, Zhang Q F. A killer-protector system regulates both hybrid sterility and segregation distortion in rice[J]. Science, 2012, 337: 1336-1340.
PMID |
| [19] | Long Y M, Zhao L F, Niu B X, Su J, Wu H, Chen Y L, Zhang Q Y, Guo J X, Zhuang C X, Mei M T, Xia J X, Wang L, Wu H B, Liu Y G. Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes[J]. Proceedings of the National Academy of Sciences, 2008, 105(48): 18871-18876. |
| [20] | Mizuta Y, Harushima Y, Kurata N. Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes[J]. Proceedings of the National Academy of Sciences, 2010, 107(47): 20417-20422. |
| [21] | Yamagata Y, Yamamoto E, Aya K, Win K T,Doi K,Sobrizal, Ito T,Kanamori H,Wu J,Matsumoto T,Matsuoka M,Ashikari M,Yoshimura A. Mitochondrial gene in the nuclear genome induces reproductive barrier in rice[J]. Proceedings of the National Academy of Sciences, 2010, 107(4): 1494-1499. |
| [22] | Xie Y Y, Niu B X, Long Y M, Li G S, Tang J T, Zhang Y L, Ren D, Liu Y G, Chen L T. Suppression or knockout of SaF/SaM overcomes the Sa-mediated hybrid male sterility in rice[J]. Journal of Integrative Plant Biology, 2017, 59(9): 669-679. |
| [23] | Koide Y, Ogino A, Yoshikawa T, Kitashima Y, Saito N, Kanaoka Y, Onishi K, Yoshitake Y, Tsukiyama T. Lineage-specific gene acquisition or loss is involved in interspecific hybrid sterility in rice[J]. Proceedings of the National Academy of Sciences, 2018, 115(9): 1955-1962. |
| [24] |
Ouyang Y D, Liu Y G, Zhang Q F. Hybrid sterility in plant: Stories from rice[J]. Current Opinion in Plant Biology, 2010, 13(2): 186-192.
PMID |
| [25] |
Ouyang Y D, Zhang Q. The molecular and evolutionary basis of reproductive isolation in plants[J]. Journal of Genetics and Genomics, 2018, 45(11): 613-620.
PMID |
| [26] | Li G W, Jin J Y, Zhou Y, Mao D, Tan C, Wang G, Ouyang Y. Genome-wide dissection of segregation distortion using multiple inter-subspecific crosses in rice[J]. Science China-Life Sciences, 2019, 62(4): 507-516. |
| [27] | Chang T T. The origin, evolution, cultivation, dissemination, and diversification of Asian and African rices[J]. Euphytica, 1976, 25(1): 425-441. |
| [28] |
Yamagishi M, Takeuchi Y, Tanaka I, Kono I, Murai K, Yano M. Segregation distortion in F2 and doubled haploid populations of temperate japonica rice[J]. Journal of Genetics, 2010, 89(2): 237.
PMID |
| [29] | Reflinur, Kim B,Jang S M,Chu S H,Bordiya Y,Akter M B,Lee J,Chin J H,Koh H J. Analysis of segregation distortion and its relationship to hybrid barriers in rice[J]. Rice, 2014, 7(1): 1-12. |
| [30] | Mackill D J. Classifying japonica rice cultivars with RAPD markers[J]. Crop Science, 1995, 35(3): 889-894. |
| [31] | Iwata N, Nagamatsu T, Omura T. Abnormal segregation of waxy and apiculus coloration by a gametophyte gene belonging to the first linkage group in rice[J]. Japanese Journal of Breeding, 1964, 14(1): 33-39. |
| [32] | Wu Y P, Ko P Y, Lee W C, Wei F J, Kuo S C, Ho S W, Hour A L, Hsing Y I, Lin Y R. Comparative analyses of linkage maps and segregation distortion of two F2 populations derived from japonica crossed with indica rice[J]. Hereditas, 2010, 147(5): 225-236. |
| [33] | 穆春华, 张发军, 李文才, 孙琦, 丁照华, 王磊, 孟昭东. 玉米叶片基因组快速提取方法研究[J]. 玉米科学, 2010, 18(3): 170-172. |
| Mu C H, Zhang F J, Li W C, Sun Q, Ding Z H, Wang L, Meng Z D. A method of genomic DNA extraction of maize[J]. Maize Science, 2010, 18(3): 170-172. (in Chinese with English abstract) | |
| [34] | 谭军, 薛庆中. 偏分离分子标记的作图方法[J]. 遗传, 2004, 26(3): 356-358. |
| Tan J, Xue Q Z. Mapping by molecular markers showing segregation distortion[J]. Hereditas(Beijing), 2004, 26(3): 356-358. (in Chinese with English abstract) | |
| [35] | Sobrizal Y. Mapping of a gene for pollen semi-sterility on chromosome 8 of rice[J]. Rice Genetic Newsletter, 2001, 18: 59-61. |
| [36] | Lyttle T W. Segregation distorters[J]. Annual Review of Genetics, 1991, 25(1): 511-581. |
| [37] |
Perfectti F. Segregation distortion of isozyme loci in cherimoya (Annona cherimola Mill)[J]. Theoretical and Applied Genetics, 1996, 93(3): 440-446.
PMID |
| [38] |
Ellstrand N C. Is gene flow the most important evolutionary force in plants?[J]. American Journal of Botany, 2014, 101(5): 737-753.
PMID |
| [39] |
Harushima Y, Kurata N, Yano M, Nagamura Y, Sasaki T, Minobe Y, Nakagahra M. Detection of segregation distortions in an indica-japonica rice cross using a high-resolution molecular map[J]. Theoretical and Applied Genetics, 1996, 92(2): 145-150.
PMID |
| [40] |
Stewart C N, Halfhill M D, Warwick S I. Transgene introgression from genetically modified crops to their wild relatives[J]. Nature Reviews Genetics, 2003, 4(10): 806-817.
PMID |
| [41] |
Gardner K A, Wittern L M, Mackay I J. A highly recombined, high-density, eight-founder wheat MAGIC map reveals extensive segregation distortion and genomic locations of introgression segments[J]. Plant Biotechnology Journal, 2016, 14(6): 1406-1417.
PMID |
| [42] | Wang L Y, He S P, Dia S, Sun G F, Liu X Y, Wang X Y, Pan Z, Jia Y H, Wang L R, Pang By, Sun X Z, Song X L, Du X M. Alien genomic introgressions enhanced fiber strength in upland cotton (Gossypium hirsutum L.)[J]. Industrial Crops and Products, 2021, 159: 113028. |
| [43] | Li W T, Liu Y, Wang J, He M, Zhou X G, Yang C, Yuan C, Wang J C, Chern M S, Yin J J, Chen W L, Ma B T, Wang Y P, Qin P, Li S G, Ronald P, Chen X W. The durably resistant rice cultivar Digu activates defence gene expression before the full maturation of Magnaporthe oryzae appressorium[J]. Molecular Plant Pathology, 2016, 17(3): 354-368. |
| [1] | 汪邑晨, 朱本顺, 周磊, 朱骏, 杨仲南. 光/温敏核不育系的不育机理及两系杂交稻的发展与展望 [J]. 中国水稻科学, 2024, 38(5): 463-474. |
| [2] | 许用强, 徐军, 奉保华, 肖晶晶, 王丹英, 曾宇翔, 符冠富. 水稻花粉管生长及其对非生物逆境胁迫的响应机理研究进展 [J]. 中国水稻科学, 2024, 38(5): 495-506. |
| [3] | 何勇, 刘耀威, 熊翔, 祝丹晨, 王爱群, 马拉娜, 王廷宝, 张健, 李建雄, 田志宏. 利用CRISPR/Cas9技术编辑OsOFP30基因创制水稻粒型突变体 [J]. 中国水稻科学, 2024, 38(5): 507-515. |
| [4] | 吕阳, 刘聪聪, 杨龙波, 曹兴岚, 王月影, 童毅, Mohamed Hazman, 钱前, 商连光, 郭龙彪. 全基因组关联分析(GWAS)鉴定水稻氮素利用效率候选基因 [J]. 中国水稻科学, 2024, 38(5): 516-524. |
| [5] | 杨好, 黄衍焱, 王剑, 易春霖, 石军, 谭楮湉, 任文芮, 王文明. 水稻中八个稻瘟病抗性基因特异分子标记的开发及应用 [J]. 中国水稻科学, 2024, 38(5): 525-534. |
| [6] | 杨铭榆, 陈志诚, 潘美清, 张汴泓, 潘睿欣, 尤林东, 陈晓艳, 唐莉娜, 黄锦文. 烟-稻轮作下减氮配施生物炭对水稻茎鞘同化物转运和产量 形成的影响 [J]. 中国水稻科学, 2024, 38(5): 555-566. |
| [7] | 熊家欢, 张义凯, 向镜, 陈惠哲, 徐一成, 王亚梁, 王志刚, 姚坚, 张玉屏. 覆膜稻田施用炭基肥对水稻产量及氮素利用的影响 [J]. 中国水稻科学, 2024, 38(5): 567-576. |
| [8] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [9] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [10] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [11] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [12] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [13] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [14] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [15] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||