
中国水稻科学 ›› 2022, Vol. 36 ›› Issue (3): 215-226.DOI: 10.16819/j.1001-7216.2022.210707
• 综述与专论 • 下一篇
王婧莹1,2, 赵广欣1,2, 邱冠凯1,2, 方军1,2,*( )
)
收稿日期:2021-07-20
修回日期:2021-09-06
出版日期:2022-05-10
发布日期:2022-05-11
通讯作者:
方军
基金资助:
WANG Jingying1,2, ZHAO Guangxin1,2, QIU Guankai1,2, FANG Jun1,2,*( )
)
Received:2021-07-20
Revised:2021-09-06
Online:2022-05-10
Published:2022-05-11
Contact:
FANG Jun
摘要:
水稻是一种广泛种植的兼性短日照植物。水稻抽穗期是直接影响产量和品种地域适应性的重要农艺性状。因此,研究该性状的影响因素并使植株在适宜的时间抽穗具有重要意义。水稻抽穗期作为一个复杂的数量性状,受内在基因网络和外界光温等条件的共同调控。目前,已经鉴定和克隆出多个控制抽穗期的关键基因,发现磷酸化和泛素化修饰在抽穗期分子机制中起重要作用。本文介绍了水稻抽穗期光周期途径的分子机制,重点阐述了磷酸化级联反应和泛素26S蛋白酶体系统对抽穗期调控的研究进展,旨在为挖掘调控抽穗期的新作用机制和改良品种地域适应性提供理论依据和实践指导。
王婧莹, 赵广欣, 邱冠凯, 方军. 水稻抽穗期途径基因的磷酸化、泛素化研究进展[J]. 中国水稻科学, 2022, 36(3): 215-226.
WANG Jingying, ZHAO Guangxin, QIU Guankai, FANG Jun. Advances in Research on the Modification of the Heading Date Genes in Rice by Phosphorylation and Ubiquitination Pathways[J]. Chinese Journal OF Rice Science, 2022, 36(3): 215-226.
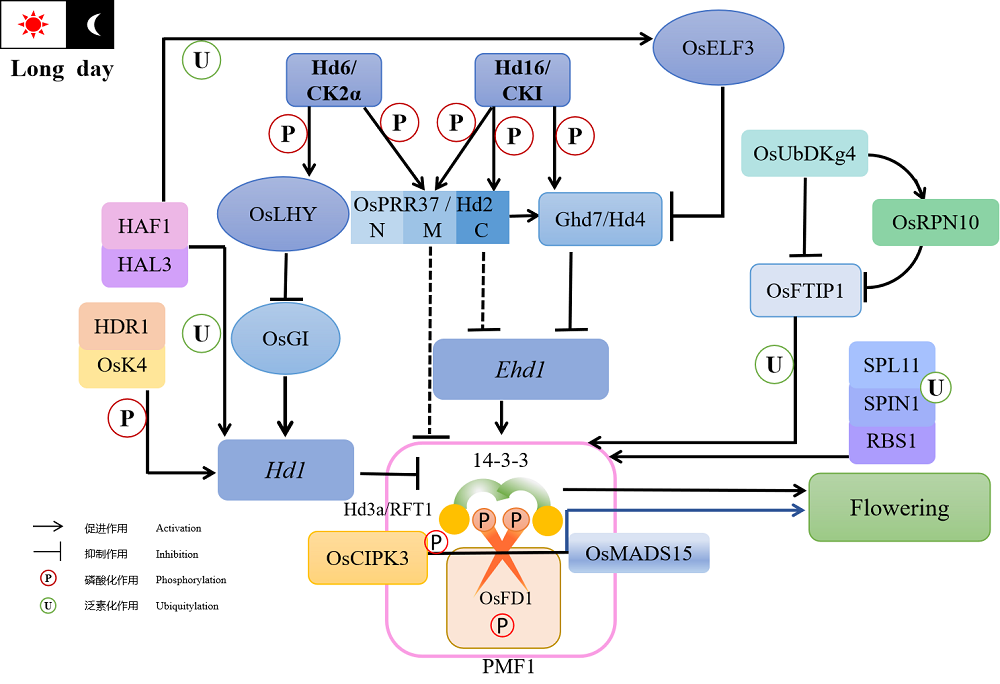
图1 磷酸化与泛素化调控水稻长日照抽穗网络 由于上述内容中仅有OsFD1和HAF1蛋白具有在短日照条件下影响抽穗期的功能,且调控途径尚不明朗,故不在图中呈现。
Fig. 1. Phosphorylation and ubiquitylation regulation network of rice flowering under long daylight condition. In the above content, only OsFD1 and HAF1 proteins have the function affecting heading date under SD conditions, and their regulatory pathways are not clear, so they are not shown in the diagram.
| [1] | Izawa T. Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice[J]. Journal of Experimental Botany, 2007, 58: 3091-3097. |
| [2] | Tsuji H, Tamaki S, Komiya R, Shimamoto K. Florigen and the photoperiodic control of flowering in rice[J]. Rice, 2008, 1: 25-35. |
| [3] | Fujino K, Sekiguchi H. Mapping of QTLs conferring extremely early heading in rice (Oryza sativa L.)[J]. Theoretical and Applied Genetics, 2005, 111: 393-398. |
| [4] | Fornara F A, de Montaigu A, Coupland G. SnapShot: Control of flowering in Arabidopsis[J]. Cell, 2010, 141: 550. |
| [5] | Masahiro Y, Takuji S. Genetic and molecular dissection of quantitative traits in rice[J]. Plant Molecular Biology, 1997, 35: 145-153. |
| [6] | Hayama R, Yokoi S, Tamaki S, Masahiro Y, Ko S. Adaptation of photoperiodic control pathways produces short-day flowering in rice[J]. Nature, 2003, 422: 719-722. |
| [7] | Kardailsky I, Shukla V K, Ahn J H, Dagenais N, Christensen S K, Nguyen J T, Chory J, Harrison M J, Weigel D. Activation tagging of the floral inducer FT[J]. Science, 1999, 286: 1962-1965. |
| [8] | Hayama R, Izawa T, Shimamoto K. Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method[J]. Plant Cell Physiology, 2002, 43: 494-504. |
| [9] | Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS[J]. Plant Cell, 2001, 12: 2473-2484. |
| [10] | Xue W Y, Xing Y Z, Weng X Y, Tang W J, Wang L, Zhou H J, Yu S B, Xu C G, Li X H, Zhang Q F. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice[J]. Nature Genetics, 2008, 40: 761-767. |
| [11] | Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A. Ehd1, a B-type response regulator in rice, confers shortday promotion of flowering and controls FT-like gene expression independently of Hd1[J]. Genes & Development, 2004, 18: 926-936. |
| [12] | Nardini M, Gnesutta N, Donati G, Gatta R, Forni C, Fossati A, Vonrhein C, Moras D, Romier C, Bolognesi M, Mantovani R. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination[J]. Cell, 2013, 152: 132-143. |
| [13] | Thirumurugan T, Ito Y, Kubo, T, Serizawa A, Kurata N. Identification, characterization and interaction of HAP family genes in rice[J]. Molecular Genetics and Genomics, 2008, 279: 279-289. |
| [14] | Petroni K, Kumimoto R W, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, Ben F, Mantovani R. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors[J]. Plant Cell, 2012, 24: 4777-4792. |
| [15] | Du A P, Tian W, Wei M G, Yan W, He H, Zhou D, Huang X, Li S G, Ouyang X H. The DTH8-Hd1 module mediates day-length-dependent regulation of rice flowering[J]. Molecular Plant, 2017, 10: 948-961. |
| [16] | Goretti D, Martignago D, Landini M, Brambilla V, Gómez-Ariza J, Gnesutta N, Galbiati F, Collani S, Takagi H, Terauchi R, Mantovani R, Fornara F. Transcriptional and post-transcriptional mechanisms limit heading date 1(Hd1) function to adapt rice to high latitudes[J]. PloS Genetics, 2017, 13: e1006530. |
| [17] | Shen C C, Liu H Y, Guan Z Y, Yan J J, Zheng T, Yan W H, Wu C Y, Zhang Q F, Yin P, Xing Y Z. Structural Insight into DNA Recognition by CCT/NF-YB/YC Complexes in Plant Photoperiodic Flowering[J]. Plant Cell, 2020, 32(11): 3469-3484. |
| [18] | Gnesutta N, Kumimoto R W, Swain S, Chiara M, Siriwardana C, Horner D S, Ben F Holt 3rd, Mantovani R. CONSTANS imparts DNA sequence specificity to the histone fold NF-YB/NF-YC dimer. Plant Cell, 2017, 29: 1516-1532. |
| [19] | 刘静, 李亚超, 周梦岩, 吴鹏飞, 马祥庆. 植物蛋白质翻译后修饰组学研究进展[J]. 生物技术通报, 2021, 37 (1): 67-76 |
| Liu J, Li Y C, Zhou M Y, Wu P F, Ma X Q. Advances in posttranslational modification of plant proteins[J]. Biotechnology Bulletin, 2021, 37(1): 67-76. | |
| [20] | Withers J, Dong X N. Post-translational regulation of plant immunity[J]. Current Opinion in Plant Biology, 2017, 38: 124-132. |
| [21] | Zhou S R, Zhu S S, Cui S, Hou H G, Wu H Q, Hao B Y, Cai L, Xu Z, Liu L L, Jiang L, Wang H Y, Wan J M. Transcriptional and post-transcriptional regulation of heading date in rice[J]. New Phytologist, 2021, 230: 943-956. |
| [22] | Millar A H, Heazlewood J L, Giglione C, Holdsworth M J, Bachmair A, Schulze W X. The scope, functions, and dynamics of posttranslational protein modifications[J]. Annual Review of Plant Biology, 2019, 70: 119-151. |
| [23] | Ruan B J, Dai P, Wang W, Sun J, Zhang W, Zhen Y, Yang J. Progress on post-translational modification of proteins[J]. Chinese Journal of Cell Biology, 2014, 7: 1027-1037 |
| [24] | Garrett R H. 生物化学. 影印版[M]. 北京: 高等教育出版社, 2005. |
| Garrett R H. Photocopy[M]. Beijing: Higher Education Press, 2005. | |
| [25] | 孙大业, 马力耕. 细胞信号转导[M]. 第2版. 北京: 科学出版社, 1998: 350-355. |
| Sun D, Ma L. Cell Signal Transduction[M]. Version 2. Beijing: Science Press, 1998: 350-355. | |
| [26] | Hanks S K, Quinn A M. Protein kinase catalytic domain sequence database: Identification of conserved features of primary structure and classification of family members[J]. Methods in Enzymology, 1991, 200: 38-62. |
| [27] | Gross S D, Anderson R A. Casein kinase: I. Spatial organization and positioning of a multifunctional protein kinase family[J]. Cell Signal, 1998, 10: 699-711. |
| [28] | Knippschild U, Gocht A, Wolff S, Huber N, Löhler J, Stöter M. The casein kinase1 family: Participation in multiple cellular processes in eukaryotes. Cell Signal, 2005, 17(6): 675-689 |
| [29] | Tan S T, Dai C, Liu H T, Xue H W. Arabidopsis casein kinase1 proteins CK1.3 and CK1.4 phosphorylate cryptochrome2 to regulate blue light signaling[J]. Plant Cell, 2013, 25: 2618-2632. |
| [30] | Daniel X, Sugano S, Tobin E M. CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis[J]. Proceedings of the National Academy of Sciences, 2004, 101: 3292-3297. |
| [31] | Ogiso E, Takahashi Y, Sasaki T, Yano M, Izawa T. The role of Casein Kinase II in flowering time regulation has diversified during evolution[J]. Plant Physiology, 2010, 152: 808-820. |
| [32] | Dai C, Xue H W. Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling[J]. The EMBO Journal, 2010, 29: 1916-1927. |
| [33] | Kwon C T, Koo B H, Kim D, Yoo S C, Paek N C. Casein Kinases I and 2α Phosphorylate Oryza sativa pseudo-response regulator 37 (OsPRR37) in photoperiodic flowering in rice[J]. Molecules and Cells, 2014: 1029-1032. |
| [34] | Murakami M, Matsushika A, Ashikari M, Yamashino T, Mizuno T. Circadian-associated rice pseudo response regulators (OsPRRs): Insight into the control of flowering time[J]. Bioscience, Biotechnology and Biochemistry, 2005, 69: 410-414. |
| [35] | Koo B H, Yoo S C, Park J W, Kwon C T, Lee B D, An G, Zhang Z Y, Li J J, Li Z Z, Paek N C. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes[J]. Molecular Plant, 2013, 6: 1877-1888. |
| [36] | Gao H, Jin M, Zheng X M, Chen J, Yuan D Y, Xin Y Y, Wang M Q, Huang D Y, Zhang Z, Zhou K N, Sheng P K, Ma J, Ma W W, Deng H F, Jiang L, Liu S J, Wang H Y, Wu C Y, Yuan L P, Wan J M. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice[J]. Proceedings of the National Academy of Sciences of USA, 2014, 111: 16337-16342. |
| [37] | Weng X Y, Wang L, Wang J, Hu Y, Du H, Xu C G, Xing Y Z, Li X H, Xiao J H, Zhang Q F. Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response[J]. Plant Physiology, 2014, 164: 735-747. |
| [38] | Huang H D, Lee T Y, Tseng S W, Horng J T. KinasePhos: A web tool for identifying protein kinase-specific phosphorylation sites[J]. Nucleic Acids Research, 2005, 33: W226-W229. |
| [39] | Li X F, Liu H Z, Wang M Q, Liu H L, Tian X J, Zhou W J, Lü T X, Wang Z Y, Chu C C, Fang J, Bu Q Y. Combinations of Hd2 and Hd4 genes determine rice adaptability to Heilongjiang Province, northern limit of China[J]. Journal of Integrative Plant Biology, 2015, 57: 698-707. |
| [40] | Cho L H, Yoon J, Pasriga R, An G. Homodimerization of Ehd1 is required to induce flowering in rice[J]. Plant Physiology, 2016, 170(4): 2159-2171. |
| [41] | Thelander M, Nilsson A, Olsson T, Johansson M, Girod P A, Schaefer D G, Zrÿd J P, Ronne H. 1 interacting proteins with homologues in vascular plants[J]. Plant Molecular Biology, 2007, 64: 559-573. |
| [42] | Sun X H, Zhang Z G, Wu J X, Cui X A, Feng D, Wang K, Xu M, Zhou L, Han X, Gu X F, Lu T G. The Oryza sativa regulator HDR1 associates with the kinase OsK4 to control photoperiodic flowering[J]. PloS Genetics, 2016, 12(3): e1005927. |
| [43] | Lu C A, Lin C C, Lee K W, Chen J L, Huang L F, Ho S L, Liu H J, Hsing Y I, Yu S M. The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice[J]. Plant Cell, 2007, 19: 2484-2499. |
| [44] | Thelander M, Olsson T, Ronne H. Snf1-related protein kinase 1 is needed for growth in a normal daynight light cycle[J]. The EMBO Journal, 2004, 23: 1900-1910. |
| [45] | Emanuelle S, Hossain M I, Moller I E, Pedersen H L, Allison M L, van de Meene, Doblin M S, Koay A, Oakhill J S, Scott J W, Willats W G T, Kemp B E, Bacic A, Gooley P R, Stapleton D I. SnRK1 from Arabidopsis thaliana is an atypical AMPK[J]. The Plant Journal, 2015, 82: 183-192. |
| [46] | Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. Hd3a and RFT1 are essential for flowering in rice[J]. Development, 2008, 135: 767-774. |
| [47] | Tamaki S,. Matsuo S, Wong H L, Yokoi S J, Shimamoto K. Hd3a protein is a mobile flowering signal in rice[J]. Science, 2007, 316: 1033-1036. |
| [48] | Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri Y A, Tamaki S, Ogaki Y, Shimada C, Nakagawa A, Kojima C, Shimamoto K. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen[J]. Nature, 2011, 476: 332-335. |
| [49] | Zhao J, Chen H Y, Ren D, Tang H W, Qiu R, Feng J L, Long Y M, Niu B X, Chen D P, Zhong T Y, Liu Y G, Guo J X. Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/ RICE FLOWERING LOCUS T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa)[J]. New Phytologist, 2015, 208(3): 936-948. |
| [50] | Peng Q, Zhu C M, Liu T, Zhang S, Feng S J, Wu C Y. Phosphorylation of OsFD1 by OsCIPK3 promotes the formation of RFT1-containing florigen activation complex for long-day flowering in rice[J]. Molecular Plant, 2021, 14(7): 1135-1148. |
| [51] | Peng Y B, Hou F X, Bai Q, Xu P Z, Liao Y X, Zhang H Y, Gu C J, Deng X S, Wu T, Chen X Q, Ali A, Wu X J. Rice calcineurin B-like protein interacting protein kinase 31 (OsCIPK31) is involved in the development of panicle apical spikelets[J]. Frontiers in Plant Science, 2018, 9: 1661. |
| [52] | 彭强. 蛋白激酶PMF1调控水稻抽穗期的分子机理研究[D]. 武汉: 华中农业大学, 2020. |
| Peng Q. Molecular mechanism of protein kinase PMF1 regulating rice heading hate in rice[D]. Wuhan: Huazhong Agricultural University, 2020. | |
| [53] | Stone S L. Role of the ubiquitin proteasome system in plant response to abiotic stress[J]. International Review of Cell and Molecular Biology, 2019, 343: 65-110. |
| [54] | Yu F F, Xie Q. Ubiquitination modification precisely modulates the ABA signaling pathway in plants[J]. Hereditas, 2017, 39(8): 692-706. |
| [55] | McClellan A J, Laugesen S H, Ellgaard L. Cellular functions and molecular mechanisms of non-lysine ubiquitination[J]. Open Biology, 2019, 9(9): 190147. |
| [56] | Dittmar G, Winklhofer K F. Linear Ubiquitin Chains: Cellular Functions and Strategies for Detection and Quantification[J]. Frontiers in Chemistry, 2019, 7: 915. |
| [57] | Fennell L M, Rahighi S, Ikeda F. Linear ubiquitin chain-binding domains[J]. The FEBS Journal, 2018, 285 (15): 2746-2761. |
| [58] | Wang F, Shi Y G. Progress in structural biology of 26S proteasome[J]. Scientia Sinica Vitae, 2014, 44: 965-974. |
| [59] | Pickart C M, Eddins M J. Ubiquitin: Structures, functions, mechanisms[J]. Biochimica et Biophysica Acta, 2004, 1695: 55-72. |
| [60] | Chen L Y, Hellmann H. Plant 3E ligases: Flexible enzymes in a sessile world[J]. Molecular Plant, 2013, 6(5): 1388-1404. |
| [61] | Vierstra R D. The ubiquitin-26S proteasome system at the nexus of plant biology[J]. Nature Reviews Molecular Cell Biology, 2009, 10: 385-397. |
| [62] | Yang Y, Fu D, Zhu C M, He Y Z, Zhang H J, Liu T, Li X H, Wu C Y. The RING-finger ubiquitin ligase HAF 1 mediates Heading date 1degradation during photoperiodic flowering in rice[J]. Plant Cell, 2015, 27: 2455-2468. |
| [63] | Stone S L, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis[J]. Plant Physiology, 2005, 137(1): 13-30. |
| [64] | Feltham R, Bettjeman B, Budhidarmo R, Mace P D, Shirley S, Condon S M, Chunduru S K, McKinlay M A, Vaux D L, Silke J, Day C L. Smac mimetics activate the E3 ligase activity of cIAP1 protein by promoting RING domain dimerization[J]. Journal of Biological Chemistry, 2011, 286: 17015-17028. |
| [65] | Zagotta M T, Hicks K A, Jacobs C I, Young J C, Hangarter R P, Meeks-Wagner D R. The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering[J]. The Plant Journal, 1996, 10: 691-702. |
| [66] | Covington M F, Panda S, Liu X L, Strayer C A, Wagner D R, Kay S A. ELF3 modulates resetting of the circadian clock in Arabidopsis[J]. Plant Cell, 2001, 13: 1305-1315. |
| [67] | Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J. GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains[J]. The EMBO Journal, 1999, 18: 4679-4688. |
| [68] | Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between thecircadian clock and the control of flowering in Arabidopsis[J]. Nature, 2001, 410(6832): 1116-1120. |
| [69] | Yang Y, Peng Q, Chen G X, Li X H, Wu C Y. OsELF3 is involved in circadian clock regulation for promoting flowering under long-day conditions in rice[J]. Molecular Plant, 2013, 6: 202-215. |
| [70] | Matsubara K, Ogiso-Tanaka E, Hori K, Ebana K, Ando T, Yano M. Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering[J]. Plant Cell Physiology, 2012, 53: 709-716. |
| [71] | Kay S A, Keith B, Shinozaki K, Chua N H. The sequence of the rice phytochrome gene[J]. Plant and Cell Physiology, 1989, 17(7): 2865-2866. |
| [72] | Sheerin D J, Menon C, Oven-Krockhaus S Z, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof Y D, Huq E, Hiltbrunner A. Light-activated Phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex[J]. Plant Cell, 2015, 27(1): 189-201. |
| [73] | Weng X Y, Wang L, Wang J, Hu Y, Du H, Xu C G, Xing Y Z, Li X H, Xiao J H, Zhang Q F. Grain Number, Plant Height, and Heading Date7is a central regulator of growth, development, and stress response[J]. Plant Physiology, 2014, 164: 735-747. |
| [74] | Zheng T H, Sun J, Zhou S R, Chen S H, Lu J, Cui S, Tian Y L, Zhang H, Cai M H, Zhu S S, Wu M M, Wang Y H, Jiang L, Zhai H Q, Wang H Y, Wan J M. Post-transcriptional regulation of Ghd7 protein stability by phytochrome and OsGI in photoperiodic control of flowering in rice[J]. New Phytologist, 2019, 224: 306-320. |
| [75] | Itoh H, Nonoue Y, Yano M, Izawa T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice[J]. Nature Genetics, 2010, 42: 635-638. |
| [76] | Andrés F, Galbraith D W, Talón M, Domingo C. Analysis of PHOTOPERIOD SENSITIVITY5 sheds light on the role of phytochromes in photoperiodic flowering in rice[J]. Plant Physiology, 2009, 151(2): 681-690. |
| [77] | Kircher S, Gil P, Kozma-Bognár L, Fejes E, Speth V, Muller T H, Bauer D, Adám E, Schäfer E, Nagy F. Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm[J]. Plant Cell, 2002, 14: 1541-1555. |
| [78] | Liu L, Liu C, Hou X L, Xi W Y, Shen L S, Tao Z, Wang Y, Yu H. FTIP1 is an essential regulator required for florigen transport[J]. PloS Biology, 2012, 10: e1001313. |
| [79] | Song S Y, Chen Y, Liu L, Wang Y W, Bao S J, Zhou X, Norman Teo Z W, Mao C Z, Gan Y B, Yu H. OsFTIP1-mediated regulation of florigen transport in rice is negatively regulated by the ubiquitin-like domain kinase OsUbDKg4[J]. Plant Cell, 2017, 29: 491-507. |
| [80] | Liu L, Zhu Y, Shen L H, Yu H. Emerging insights into florigen transport[J]. Plant Biology, 2013, 16: 607-613. |
| [81] | Galvão R, Kota U, Soderblom E, Goshe M, Boss W. Characterization of a new family of protein kinases from Arabidopsis containing phosphoinositide 3/4-kinase and ubiquitin-like domains[J]. Biochemical Journal, 2008, 409: 117-127. |
| [82] | Balla A, Balla T. Phosphatidylinositol 4-kinases: Old enzymes with emerging functions[J]. Trends in Cell Biology, 2006, 16(7): 351-361. |
| [83] | Fruman D A, Meyers R E, Cantley L C. Phosphoinositide kinases[J]. Annual Review of Biochemistry, 1998, 67: 481-507. |
| [84] | Lucas J I, Arnau V, Marin I. Comparative genomics and protein domain graph analyses link ubiquitination and RNA metabolism[J]. Molecular Biology, 357(1): 9-17. |
| [85] | Liu J L, Li W, Ning Y S, Shirsekar G, Cai Y H, Wang X L, Dai L Y, Wang Z L, Liu W D, Wang G L. The U-box E3 ligase SPL11/PUB13 is a convergence point of defense and flowering signaling in plants[J]. Plant Physiology, 2012, 160(1): 28-37. |
| [86] | Zeng L R, Park C H, Venu R C, Gough J, Wang G L. Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins[J]. Molecular Plant, 2008, 1(5): 800-815. |
| [87] | Vernet C, Artzt K. STAR, a gene family involved in signal transduction and activation of RNA[J]. Trends in Genetics, 1997, 13(12): 479-484. |
| [88] | Vega-Sa´nchez M E, Zeng L, Chen S B, Leung H, Wang G L. SPIN1, a K homology domain protein negatively regulated and ubiquitinated by the E3 ubiquitin ligase SPL11, is involved in flowering time control in rice[J]. Plant Cell, 2008, 20(6): 1456-1469. |
| [89] | Lorković Z J, Barta A. A Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana[J]. Nucleic Acids Research, 2002, 30: 623-635. |
| [90] | Cai Y H, Vega-Sa´nchez M E, Park C H, Bellizzi M, Guo Z J, Wang G L. RBS1, an RNA binding protein, interacts with SPIN1 and is involved in flowering time control in rice[J]. PLoS ONE, 2014, 9(1): e87258. |
| [91] | Sun C H, Chen D, Fang J, Wang P G, Deng X J, Chu C C. Understanding the genetic and epigenetic architecture in complex network of rice flowering pathways[J]. Protein Cell, 2014, 5(12): 889-898. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||