
中国水稻科学 ›› 2021, Vol. 35 ›› Issue (6): 629-638.DOI: 10.16819/j.1001-7216.2021.210206
• • 上一篇
曹煜东, 肖湘谊, 叶乃忠, 丁晓雯, 易晓璇, 刘金灵*( ), 肖应辉*(
), 肖应辉*( )
)
收稿日期:2021-02-19
修回日期:2021-03-16
出版日期:2021-11-10
发布日期:2021-11-10
通讯作者:
刘金灵,肖应辉
基金资助:
Yudong CAO, Xiangyi XIAO, Naizhong YE, Xiaowen DING, Xiaoxuan YI, Jinling LIU*( ), Yinghui XIAO*(
), Yinghui XIAO*( )
)
Received:2021-02-19
Revised:2021-03-16
Online:2021-11-10
Published:2021-11-10
Contact:
Jinling LIU, Yinghui XIAO
摘要:
【目的】水稻粒形为多基因控制的复杂数量性状,同时影响稻谷产量和稻米外观及碾磨品质,挖掘粒形相关基因并解析其遗传机制,对于水稻高产和优质品种选育具有重要意义。【方法】以前期利用大粒籼稻品种特大籼(TDX)为供体亲本、小粒粳稻品种日本晴(Nipponbare,NPB)为轮回亲本获得的一个大粒近等基因系,在水稻第2染色体上初定位粒形调控基因GS2.2(grain size 2.2)的基础上,对GS2.2基因进行了精细定位和候选功能基因的克隆鉴定。【结果】利用BC4F2群体中2887份极小粒单株及该群体中70份基因型杂合的大粒单株自交获得的BC4F3群体,将GS2.2基因精细定位在2M-7-1与2M-9之间的160.6 kb的区间内。该区间编码18个基因开放阅读框(ORF1-18)。测序发现ORF18基因在大粒近等基因系与小粒日本晴等位基因间存在5个SNP差异,ORF18编码生长素调控因子蛋白OsGRF4。采用CRISPR/Cas9基因编辑技术对大粒近等基因系中ORF18进行敲除,发现ORF18敲除株系粒长变短,证实ORF18是GS2.2的功能基因。进一步对ORF18大粒近等基因系和基因编辑敲除株系进行稻瘟菌室内离体和病圃接种鉴定,发现ORF18大粒近等基因系稻瘟病抗性增强,而基因编辑敲除株系稻瘟病抗性减弱,表明ORF18正调控水稻稻瘟病抗性。【结论】本研究发现的生长素调控因子OsGRF4协同调控水稻粒形和稻瘟病抗性,为协调水稻高产和抗性育种提供了重要的基因资源。
曹煜东, 肖湘谊, 叶乃忠, 丁晓雯, 易晓璇, 刘金灵, 肖应辉. 生长素调控因子OsGRF4协同调控水稻粒形和稻瘟病抗性[J]. 中国水稻科学, 2021, 35(6): 629-638.
Yudong CAO, Xiangyi XIAO, Naizhong YE, Xiaowen DING, Xiaoxuan YI, Jinling LIU, Yinghui XIAO. Auxin Regulator OsGRF4 Simultaneously Regulates Rice Grain Shape and Blast Resistance[J]. Chinese Journal OF Rice Science, 2021, 35(6): 629-638.
| 引物用途 Primer usage | 引物名称 Primer name | 引物序列 Primer sequence |
|---|---|---|
| CRISPR/Cas9载体构建 Constriction of CRISPR/Cas9 vector | CRS-GRF4-F | GCCGCAGCTCCTCGTACTGCGCCG |
| CRS-GRF4-R | AAACCGGCGCAGTACGAGGAGCTG | |
| 敲除转基因植株鉴定 Identification of knockout transgenic plants | GRF4-CRSJD-F | AACCCATTTTCTTGGCTC |
| GRF4-CRSJD-R | CGCCTGATCGGAATAAAC |
表1 CRISPR/Cas9载体构建和基因敲除植株鉴定所用引物
Table 1 Primers for construction and identification of CRISPR/Cas9 vectors.
| 引物用途 Primer usage | 引物名称 Primer name | 引物序列 Primer sequence |
|---|---|---|
| CRISPR/Cas9载体构建 Constriction of CRISPR/Cas9 vector | CRS-GRF4-F | GCCGCAGCTCCTCGTACTGCGCCG |
| CRS-GRF4-R | AAACCGGCGCAGTACGAGGAGCTG | |
| 敲除转基因植株鉴定 Identification of knockout transgenic plants | GRF4-CRSJD-F | AACCCATTTTCTTGGCTC |
| GRF4-CRSJD-R | CGCCTGATCGGAATAAAC |
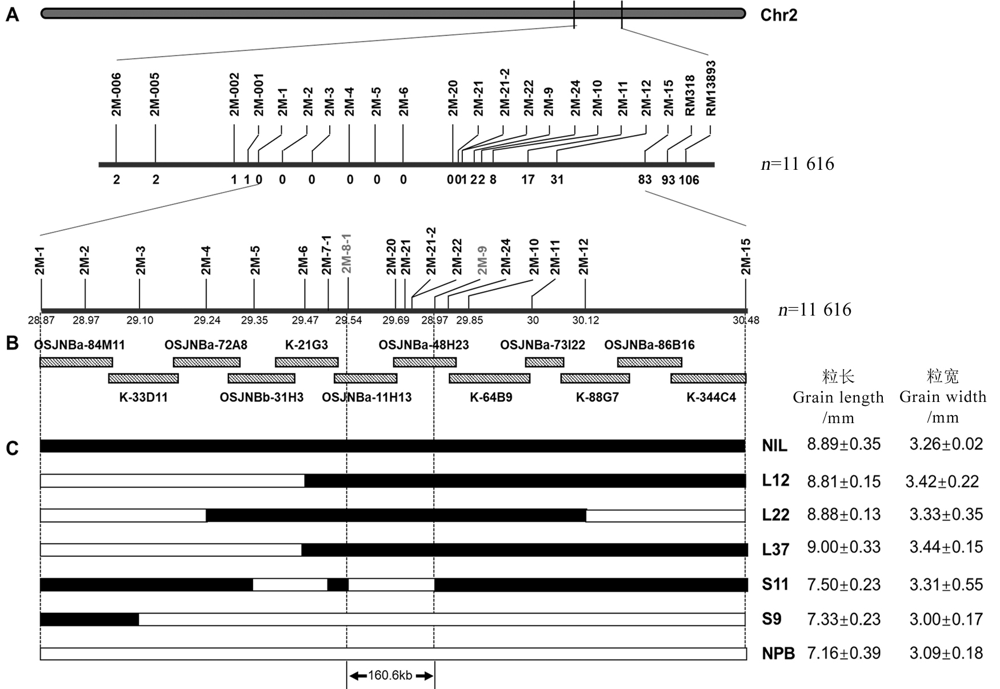
图1 GS2.2基因的精细定位与物理图谱构建 A-GS2.2精细定位遗传图谱;B-GS2.2位点BAC克隆重叠群和物理图谱;C-重组株系迭代法确定GS2.2物理区间,其中NIL为大粒近等基因系,NPB为日本晴,“L+数字”代表表型为大粒的重组株系,“S+数字”代表表型为小粒的重组株系,右侧为对应株系粒长和粒宽。
Fig. 1. Construction of genetic map and physical map of GS2.2. A, Genetic map of GS2.2; B, BAC clone contig and physical map of GS2.2 locus; C, The GS2.2 physical interval was determined by overlapping recombinant lines. NIL, Near-isogenic line with large grains; NPB, Nipponbare; “L+Number” represents a recombinant strain with large grains, and the “S+Number” represents a recombinant strain with small grains. The phenotype of grain length and width for NPB, NIL and recombinant lines was presented on the right.
| 候选基因 Candidate gene | 蛋白编码结构 Protein coding structure | |
|---|---|---|
| ORF1 | Probable WRKY transcription factor 14 | |
| ORF2 | Protein NRT1/ PTR FAMILY 8.3 | |
| ORF3 | ADP-ribosylation factor | |
| ORF4 | Uncharacterized LOC4330423 | |
| ORF5 | Putative surface protein SACOL0050 | |
| ORF6 | 60S ribosomal protein L12-1 | |
| ORF7 | DNA topoisomerase 2 | |
| ORF8 | Protein-coding | |
| ORF9 | Os02g0699800 | |
| ORF10 | Transcription initiation factor TFIID subunit 8 | |
| ORF11 | Uncharacterized protein At1g51745 | |
| ORF12 | Cell division cycle 20.2, cofactor of APC complex | |
| ORF13 | Myb family transcription factor APL | |
| ORF14 | Ubiquinol oxidase 2, mitochondrial | |
| ORF15 | Polyamine transporter PUT1 | |
| ORF16 | Cyclin-dependent kinase F-4 | |
| ORF17 | Protein Brevis radix-like 2 | |
| ORF18 | Growth-regulating factor 4 | |
表2 GS2.2候选基因预测分析
Table 2 Prediction and analysis of GS2.2 candidate genes.
| 候选基因 Candidate gene | 蛋白编码结构 Protein coding structure | |
|---|---|---|
| ORF1 | Probable WRKY transcription factor 14 | |
| ORF2 | Protein NRT1/ PTR FAMILY 8.3 | |
| ORF3 | ADP-ribosylation factor | |
| ORF4 | Uncharacterized LOC4330423 | |
| ORF5 | Putative surface protein SACOL0050 | |
| ORF6 | 60S ribosomal protein L12-1 | |
| ORF7 | DNA topoisomerase 2 | |
| ORF8 | Protein-coding | |
| ORF9 | Os02g0699800 | |
| ORF10 | Transcription initiation factor TFIID subunit 8 | |
| ORF11 | Uncharacterized protein At1g51745 | |
| ORF12 | Cell division cycle 20.2, cofactor of APC complex | |
| ORF13 | Myb family transcription factor APL | |
| ORF14 | Ubiquinol oxidase 2, mitochondrial | |
| ORF15 | Polyamine transporter PUT1 | |
| ORF16 | Cyclin-dependent kinase F-4 | |
| ORF17 | Protein Brevis radix-like 2 | |
| ORF18 | Growth-regulating factor 4 | |
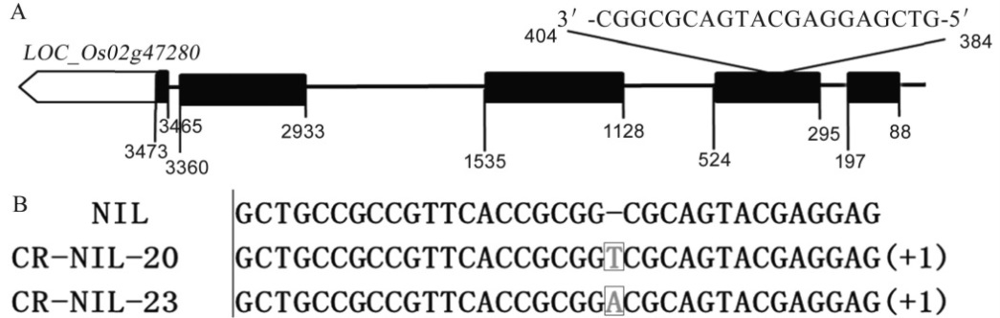
图3 OsGRF4 CRISPR/Cas9基因编辑靶点序列和编辑株系基因型 A―OsGRF4基因编辑靶点序列;B―OsGRF4基因编辑株系敲除系靶点突变基因型。
Fig. 3. Sequence of target site for OsGRF4 CRISPR / Cas9 gene editing and genotype of OsGRF4 CRISPR / Cas9 gene editing lines in NIL background. A, Sequence of target site for OsGRF4 CRISPR / Cas9 gene editing; B, Genotype of OsGRF4 CRISPR / Cas9 gene editing lines in NIL.
| 转基因株系 Transgenic lines | 基因型 Genotype | 基因编辑类型 Gene editing type |
|---|---|---|
| Reference | GCTGCCGCCGTTCACCGCGGCGCAGTACGAGGAGC | |
| CR-NIL-15 | GCTGCCGCCGTTCACCGCGGTCGCAGTACGAGGAGC | +1 |
| CR-NIL-16 | GCTGCCGCCGTTCACCGCGGACGCAGTACGAGGAG | +1 |
| GCTGCCGCCGTTCACCGCGGCCGCAGTACGAGGAG | +1 | |
| CR-NIL-17 | AGGCTGCCGCCGTTCACCGC-(46 bp del)CTGGTGGCAGGCGTG | -46 |
| AGGCTGCCGCCGTTCACCGCGGGCGCAGTACGAGG | +1 | |
| CR-NIL-19 | GGCTGCCGCCGTTCACCGCGGCGCAGTACGAGGAG (WT) | +0 |
| GGCTGCCGCCGTTCACCGCGTACGCAGTACGAGGA | +1 | |
| CR-NIL-20 | GCTGCCGCCGTTCACCGCGGTCGCAGTACGAGGAG | +1 |
| CR-NIL-23 | GCTGCCGCCGTTCACCGCGGACGCAGTACGAGGAG | +1 |
| CR-NIL-22 | GCTGCCGCCGTTCACCGCGGGCGCAGTACGAGGAG | +1 |
| GCTGCCGCCGTTCACCGCGGTCGCAGTACGAGGAG | +1 |
表3 OsGRF4 CRISPR/Cas9基因编辑敲除突变类型
Table 3 OsGRF4 CRISPR / Cas9 gene editing knockout mutation types.
| 转基因株系 Transgenic lines | 基因型 Genotype | 基因编辑类型 Gene editing type |
|---|---|---|
| Reference | GCTGCCGCCGTTCACCGCGGCGCAGTACGAGGAGC | |
| CR-NIL-15 | GCTGCCGCCGTTCACCGCGGTCGCAGTACGAGGAGC | +1 |
| CR-NIL-16 | GCTGCCGCCGTTCACCGCGGACGCAGTACGAGGAG | +1 |
| GCTGCCGCCGTTCACCGCGGCCGCAGTACGAGGAG | +1 | |
| CR-NIL-17 | AGGCTGCCGCCGTTCACCGC-(46 bp del)CTGGTGGCAGGCGTG | -46 |
| AGGCTGCCGCCGTTCACCGCGGGCGCAGTACGAGG | +1 | |
| CR-NIL-19 | GGCTGCCGCCGTTCACCGCGGCGCAGTACGAGGAG (WT) | +0 |
| GGCTGCCGCCGTTCACCGCGTACGCAGTACGAGGA | +1 | |
| CR-NIL-20 | GCTGCCGCCGTTCACCGCGGTCGCAGTACGAGGAG | +1 |
| CR-NIL-23 | GCTGCCGCCGTTCACCGCGGACGCAGTACGAGGAG | +1 |
| CR-NIL-22 | GCTGCCGCCGTTCACCGCGGGCGCAGTACGAGGAG | +1 |
| GCTGCCGCCGTTCACCGCGGTCGCAGTACGAGGAG | +1 |
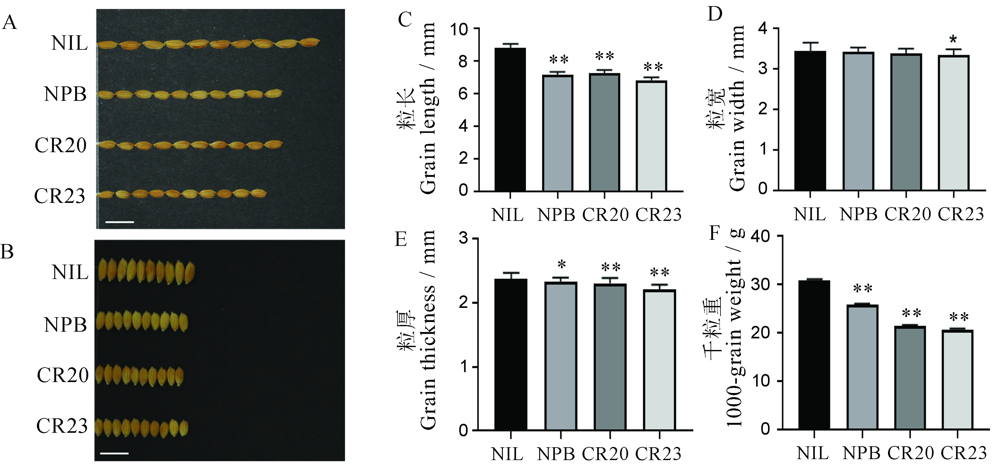
图4 GS2.2-NIL背景下OsGRF4基因编辑转基因水稻的粒形表型 A和B―GS2.2-NIL背景下OsGRF4基因编辑转基因株系粒长和粒宽;C~F―GS2.2-NIL背景下OsGRF4基因编辑转基因株系粒长、粒宽、粒厚和千粒重统计。*P<0.05; **P<0.01(t测验)。误差线表示SD (n=30)。NIL―GS2.2-NIL; CR20-CR-NIL-20; CR23-CR-NIL-23.
Fig. 4. Grain shape of gene editing lines in GS2.2-NIL background. A and B, Phenotype of grain length and width. C-F, Statistic data of grain length, width, thickness and 1000-grain weight. *P<0.05; **P<0.01(t-test). Error bar indicated the SD value (n=30). NIL, GS2.2-NIL; CR20, CR-NIL-20; CR23, CR-NIL-23.
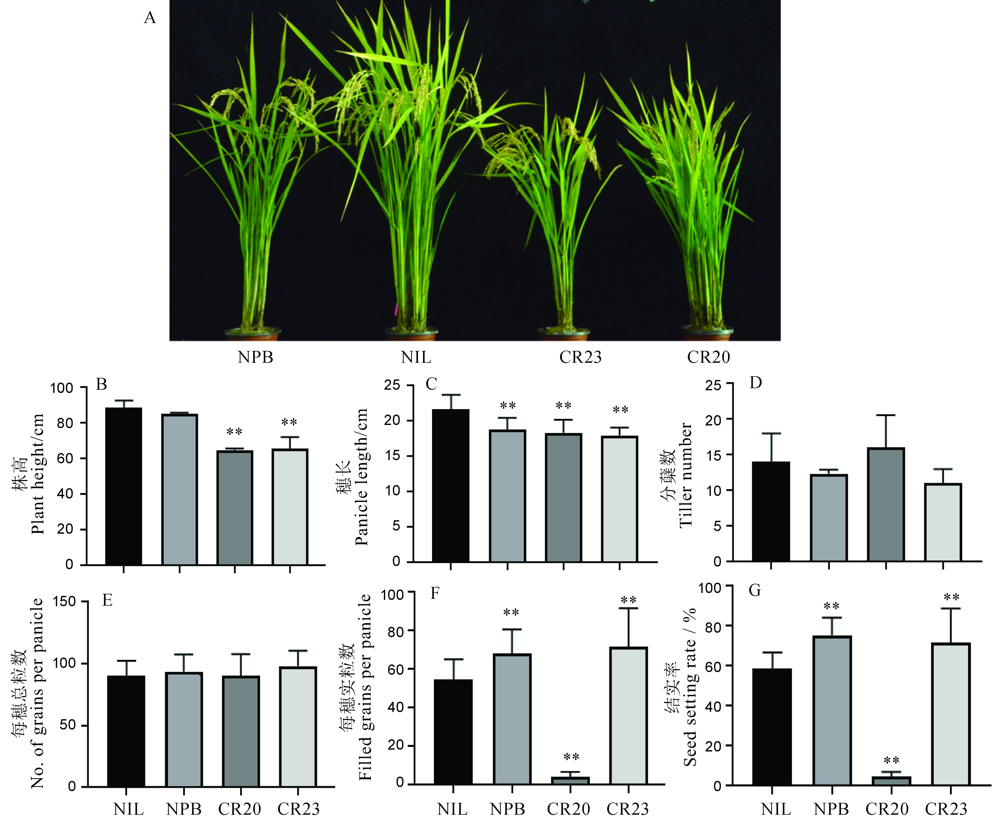
图5 NIL背景下OsGRF4基因编辑株系农艺性状表型 A―NIL背景下OsGRF4基因编辑株系株型;B~G, NIL背景下OsGRF4基因编辑株系株高、穗长、分蘖数、每穗总粒数、每穗实粒数和结实率。显著性分析采用t测验,图中误差值以SD表示(n=3)。NIL―GS2.2-NIL; CR20-CR-NIL-20; CR23-CR-NIL-23.
Fig. 5. Phenotype of agronomic traits of OsGRF4 gene editing lines in NIL background. A, Plant architecture for gene editing lines. B-G, Plant height, panicle length, tiller number, total grains per panicle, number of full grains per panicle and seed setting rate for gene editing lines. The t-test was used for significance analysis. Error bar indicated the SD value (n=3). **P<0.01.NIL, GS2.2-NIL; CR20, CR-NIL-20; CR23, CR-NIL-23.
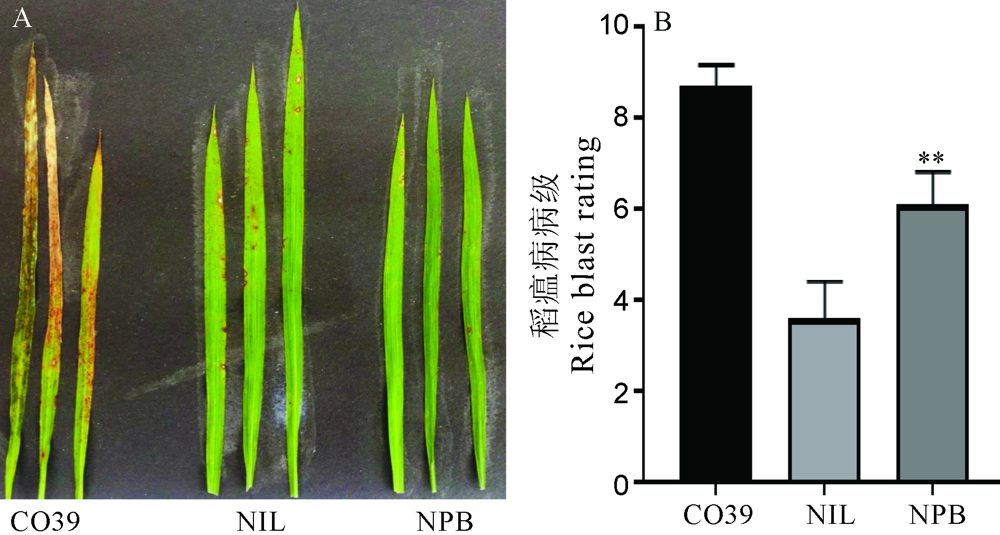
图6 GS2.2-NIL和日本晴(NPB)在自然病圃发病症状 A―叶片病圃发病症状;B―叶片病圃发病级别统计。显著性分析采用t测验,图中误差值以SD表示(n=10)。**P<0.01(t测验)。
Fig. 6. Symptoms of GS2.2-NIL and Nipponbare(NPB) after infected by rice blast fungal in natural disease nurse. A, Symptoms of leaf after rice blast fungal infection in the natural disease nurse; B, Leaf blast rating after rice blast fungal infection in the natural disease nurse. The t-test was used for significance analysis. Error bar indicated the SD(n=10). **P<0.01。
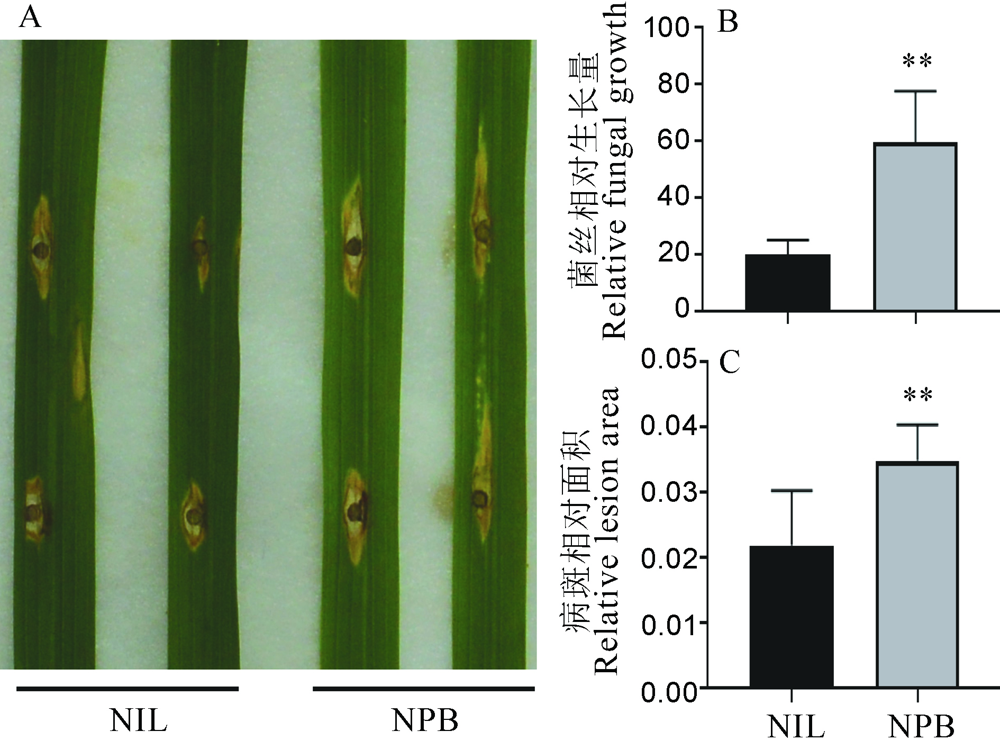
图7 GS2.2-NIL和日本晴(NPB)稻瘟病离体接种发病症状 A―病斑症状;B―病斑相对菌丝量生物量;C―病斑相对面积。显著性分析采用t测验,图中误差值示SD(n=3)。**P<0.01(t测验)。
Fig. 7. Symptoms of GS2.2-NIL and Nipponbare(NPB) after in vitro inoculation with rice blast fungus. A, Leaf disease phenotype; B, Relative blast fungal biomass in the infected leaf; C, Area of leaf infected by blast fungal. The t-test was used for significance analysis, error bar indicated the SD value(n=3). **P<0.01(t-test).
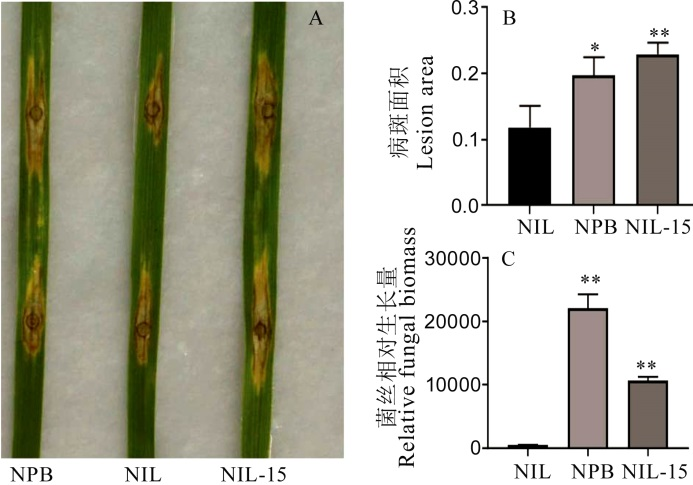
图8 日本晴、GS2.2-NIL和GS2.2-NIL敲除株系稻瘟菌接种发病症状 A―病斑表型;B―病斑面积;C―病斑相对菌丝量生物量。显著性分析采用t测验, 图中误差线示SD(n=3)。*P<0.05; **P<0.01. NIL-15, CR-NIL-15.
Fig. 8. Symptoms of GS2.2-NIL, Nipponbare(NPB) and CRISPR/Cas9 editing lines after in vitro inoculation with rice blast fungus. A, Phenotype of infection; B, Infected leaf area; C, The relative fungal biomass. The t-test was used for significance analysis, error bar indicated the SD value (n=3). *P<0.05; **P<0.01(t-test). NIL-15, CR-NIL-15.
| [1] | 康艺维, 陈玉宇, 张迎信. 水稻粒型基因克隆研究进展及育种应用展望[J]. 中国水稻科学, 2020, 34(6): 479-490. |
| Kang Y W, Chen Y Y, Zhang Y X.Research progress and breeding prospects of grain size associated genes in rice[J]. Chinese Journal of Rice Science, 2020, 34(6): 479-490. (in Chinese with English abstract) | |
| [2] | 杨维丰, 詹鹏麟, 林少俊, 苟亚军, 张桂权, 王少奎. 水稻粒形的遗传研究进展[J]. 华南农业大学学报, 2019, 40(5): 203-210. |
| Yang W F, Zhan P L, Lin S J, Gou Y J, Zhang G Q, Wang S K.Research progress of grain shape genetics in rice[J]. Journal of South China Agricultural University, 2019, 40(5): 203-210. (in Chinese with English abstract) | |
| [3] | 刘喜, 牟昌铃, 周春雷, 程治军, 江玲, 万建民. 水稻粒型基因克隆和调控机制研究进展[J]. 中国水稻科学, 2018, 32(1): 1-11. |
| Liu X, Mou C L, Zhou C L, Cheng Z J, Jiang L, Wan J M.Research progress on cloning and regulation mechanism of rice grain shape genes[J]. Chinese Journal of Rice Science, 2018, 32(1): 1-11. (in Chinese with English abstract) | |
| [4] | Huang R, Jiang L, Zheng J, Wang T, Wang H, Huang Y, Hong Z.Genetic bases of rice grain shape: So many genes, so little known[J]. Trends in Plant Science, 2013, 18(4): 218-226. |
| [5] | 尉鑫, 曾智锋, 杨维丰, 韩婧, 柯善文. 水稻粒形遗传调控研究进展[J]. 安徽农业科学, 2019, 47(5): 21-28. |
| Wei X, Zeng Z F, Yang W F, Han J, Ke S W.Advances in studies on genetic regulation of rice grain shape.Journal of Anhui Agricultural Science, 2019, 47(5): 21-28. (in Chinese with English abstract) | |
| [6] | 马超, 原佳乐, 张苏, 贾琦石, 冯雅岚. GRF转录因子对植物生长发育及胁迫响应调控的分子机制[J]. 核农学报, 2017, 31(11): 2145-2153. |
| Ma C, Yuan J L, Zhang S, Jia Q S, Feng Y L.The molecular mechanisms of growth-regulating factors (GRFs) in plant growth, development and stress response[J]. Journal of Nuclear Agricultural Sciences, 2017, 31(11): 2145-2153. (in Chinese with English abstract) | |
| [7] | 袁岐, 张春利, 赵婷婷, 许向阳. 植物中GRF转录因子的研究进展[J]. 基因组学与应用生物学, 2017, 36(8): 3145-3151. |
| Yuan Q, Zhang C L, Zhao T T, Xu X Y.Research advances of GRF transcription factor in plant[J]. Genomics and Applied Biology, 2017, 36(8): 3145-3151. (in Chinese with English abstract) | |
| [8] | Szczygieł-Sommer A, Gaj M D.The miR396-GRF regulatory module controls the embryogenic response in Arabidopsis via an auxin-related pathway[J]. Inter- national Journal of Molecular Sciences, 2019, 20: 5221. |
| [9] | Liang G, He H, Li Y, Wang F, Yu D.Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis[J]. Plant Physiology, 2014, 164(1): 249-258. |
| [10] | van Knaap E, Kim J H, Kende H. A novel gibberellin- induced gene from rice and its potential regulatory role in stem growth[J]. Plant Physiology, 2000, 122(3): 695. |
| [11] | Chandran V, Wang H, Gao F, Cao X L, Chen Y P, Li G B, Zhu Yong, Yang X M, Zhang L L, Zhao Z X, Zhao J H, Wang Y G, Li S C, Fan J, Li Y, Zhao J Q, Li S Q, Wang W M. miR396-OsGRFs module balances growth and rice blast disease-resistance[J]. Frontiers in Plant Science, 2019, 9: 1999. |
| [12] | Dai Z, Tan J, Zhou C, Yang X, Yang F, Zhang S, Sun S, Miao X, Shi Z.The OsmiR396-OsGRF8-OsF3H- flavonoid pathway mediates resistance to the brown planthopper in rice (Oryza sativa)[J]. Plant Biotechnology Journal, 2019, 17: 1657-1669. |
| [13] | Sun P, Zhang W, Wang Y, He Q, Shu F, Liu H, Wang J, Wang J, Yuan L, Deng H.OsGRF4 controls grain shape, panicle length and seed shattering in rice[J]. Journal of Integrative Plant Biology, 2016, 58(10): 836-847. |
| [14] | Li S, Gao F, Xie K, Zeng X, Cao Y, Zeng J, He Z, Ren Y, Li W, Deng Q, Wang S, Zheng A, Zhu J, Liu H, Wang L, Li P.The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice[J]. Plant Biotechnology Journal, 2016, 14(11): 2134-2146. |
| [15] | Li S, Tian Y, Wu K, Ye Y, Yu J, Zhang J, Liu Q, Hu M, Li H, Tong Y, Harberd N P, Fu X.Modulating plant growth-metabolism coordination for sustainable agriculture[J]. Nature, 2018, 560(7720): 595-600. |
| [16] | Chen X, Jiang L, Zheng J, Chen F, Wang T, Wang M, Tao Y, Wang H, Hong Z, Huang Y, Huang R.A missense mutation in Large Grain Size 1 increases grain size and enhances cold tolerance in rice[J]. Journal of Experimental Botany, 2019, 70(15): 3851-3866. |
| [17] | 叶乃忠. 水稻粒形基因GS2.2的精细定位[D]. 长沙: 湖南农业大学, 2016. |
| Ye N Z.Fine mapping of rice grain shape gene GS2.2[D]. Changsha: Hunan Agricultural University, 2016. (in Chinese with English abstract) | |
| [18] | 楼巧君, 陈亮, 罗利军. 三种水稻基因组DNA快速提取方法的比较[J]. 分子植物育种, 2005, 13(5): 749-752. |
| Lou Q J, Chen L, Luo L J.Comparison of three rapid rice DNA extraction methods[J]. Molecular Plant Breeding, 2005, 13(5): 749-752. (in Chinese with English abstract) | |
| [19] | 张会军. 水稻Pi21和OsBadh2基因编辑改良空育131的稻瘟病抗性及香味品质[D]. 武汉: 华中农业大学, 2016. |
| Zhang H J.Rice blast resistance and aroma quality improved by editing rice Pi21 and OsBadh2 genes [D]. Wuhan: Huazhong Agricultural University, 2016. (in Chinese with English abstract) | |
| [20] | 王文文, 孙红正, 李俊周, 彭廷, 杜彦修, 张静, 赵全志. 水稻miR395d基因过表达载体的构建及功能初步分析[J]. 分子植物育种, 2019, 17(9): 2876-2881. |
| Wang W W, Sun H Z, Li J Z, Peng T, Du Y X, Zhang J, Zhao Q Z.Construction and preliminary functional analysis of over-expression vector of mir395d gene in rice[J]. Molecular Plant Breeding, 2019, 17(9): 2876-2881. (in Chinese with English abstract) | |
| [21] | 崔华威, 杨艳丽, 黎敬涛, 罗文富, 苗爱敏, 胡振兴, 韩小女. 一种基于Photoshop的叶片相对病斑面积快速测定方法[J]. 安徽农业科学, 2009, 37(22): 10760-10762, 10805. |
| Cui H W, Yang Y L, Li J T, Luo W F, Miao A M, Hu Z X, Han X N.A faster method for measuring relative lesion area on leaves based on software photoshop[J]. Journal of Anhui Agricultural Science, 2009, 37(22): 10760-10762, 10805. (in Chinese with English abstract) | |
| [22] | International Rice Research Institute. Standard Evaluation System for Rice (SES)[M]. Los Baños, Philippines: International Rice Research Institute, 1996: 17-18. |
| [23] | 孙平勇, 张武汉, 张莉, 舒服, 何强, 彭志荣, 邓华凤. 水稻氮高效、耐冷基因OsGRF4功能标记的开发及其利用[J]. 作物学报, 2021, 47(4): 684-690. |
| Sun P Y, Zhang W H, Zhang L, Shu F, He Q, Peng Z R, Deng H F.Development and application of functional marker for high nitrogen use efficiency and chilling tolerance gene OsGRF4 in rice. Acta Agronomica Sinica, 2021, 47(4): 684-690. (in Chinese with English abstract) | |
| [24] | 刘学英, 李姗, 吴昆, 刘倩, 高秀华, 傅向东. 提高农作物氮肥利用效率的关键基因发掘与应用[J]. 科学通报, 2019, 64(25): 2633-2640. |
| Liu X Y, Li S, Wu K, Liu Q, Gao X H, Fu X D.Sustainable crop yields from the coordinated modulation of plant growth and nitrogen metabolism[J]. Chinese Science Bulletin, 2019, 64(25): 2633-2640. (in Chinese with English summary) | |
| [25] | Li W, Zhu Z, Chen M, Yin J, Yang C, Ran L, Cheng M, He M, Wang K, Wang J, Zhou X, Zhu X, Chen Z, Wang J, Zhao W, Ma B, Qin P, Chen W, Wang Y, Liu J, Wu X, Li P, Wang J, Zhu L, Li S, Chen X. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance [J]. Cell, 2017, 170(1): 114-126.e15. |
| [26] | Zhou X, Liao H, Chen M, Yin J, Chen Yi, Wang J, Zhu X, Chen Z, Yuan C, Wang W, Wu X, Li P, Zhu L, Li S, Ronald P C, Chen X.Loss of function of a rice TPR-domain RNA-binding protein confers broad- spectrum disease resistance[J]. Proceedings of the National Academy of Sciences of the Unite States of America, 2018, 115: 3174-3179. |
| [27] | Wang J, Zhou L, Shi H, Chen M, Yu H, Yi H, He M, Yin J, Zhu X, Li Y, Li W, Liu J, Wang J, Chen X, Qing H, Wang Y, Liu G, Wang W, Li P, Wu X, Zhu L, Zhou J, Ronald P C, Li S, Li J, Chen X,.A single transcription factor promotes both yield and immunity in rice[J]. Science, 2018, 361: 1026-1028. |
| [1] | 汪邑晨, 朱本顺, 周磊, 朱骏, 杨仲南. 光/温敏核不育系的不育机理及两系杂交稻的发展与展望 [J]. 中国水稻科学, 2024, 38(5): 463-474. |
| [2] | 许用强, 徐军, 奉保华, 肖晶晶, 王丹英, 曾宇翔, 符冠富. 水稻花粉管生长及其对非生物逆境胁迫的响应机理研究进展 [J]. 中国水稻科学, 2024, 38(5): 495-506. |
| [3] | 何勇, 刘耀威, 熊翔, 祝丹晨, 王爱群, 马拉娜, 王廷宝, 张健, 李建雄, 田志宏. 利用CRISPR/Cas9技术编辑OsOFP30基因创制水稻粒型突变体 [J]. 中国水稻科学, 2024, 38(5): 507-515. |
| [4] | 吕阳, 刘聪聪, 杨龙波, 曹兴岚, 王月影, 童毅, Mohamed Hazman, 钱前, 商连光, 郭龙彪. 全基因组关联分析(GWAS)鉴定水稻氮素利用效率候选基因 [J]. 中国水稻科学, 2024, 38(5): 516-524. |
| [5] | 杨好, 黄衍焱, 王剑, 易春霖, 石军, 谭楮湉, 任文芮, 王文明. 水稻中八个稻瘟病抗性基因特异分子标记的开发及应用 [J]. 中国水稻科学, 2024, 38(5): 525-534. |
| [6] | 杨铭榆, 陈志诚, 潘美清, 张汴泓, 潘睿欣, 尤林东, 陈晓艳, 唐莉娜, 黄锦文. 烟-稻轮作下减氮配施生物炭对水稻茎鞘同化物转运和产量 形成的影响 [J]. 中国水稻科学, 2024, 38(5): 555-566. |
| [7] | 熊家欢, 张义凯, 向镜, 陈惠哲, 徐一成, 王亚梁, 王志刚, 姚坚, 张玉屏. 覆膜稻田施用炭基肥对水稻产量及氮素利用的影响 [J]. 中国水稻科学, 2024, 38(5): 567-576. |
| [8] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [9] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [10] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [11] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [12] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [13] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [14] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [15] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||