
中国水稻科学 ›› 2020, Vol. 34 ›› Issue (3): 278-286.DOI: 10.16819/j.1001-7216.2020.9056
• 实验技术 • 上一篇
陈铭学1,2,#, 杨欢2,#, 曹赵云2, 马有宁2, 曹珍珍2, 程方民1,*( )
)
收稿日期:2019-05-15
修回日期:2019-08-01
出版日期:2020-05-15
发布日期:2020-05-10
通讯作者:
程方民
作者简介:#共同第一作者
基金资助:
Mingxue CHEN1,2,#, Huan YANG2,#, Zhaoyun CAO2, Youning MA2, Zhenzhen CAO2, Fangmin CHENG1,*( )
)
Received:2019-05-15
Revised:2019-08-01
Online:2020-05-15
Published:2020-05-10
Contact:
Fangmin CHENG
About author:#These authors contributed equally to this work
摘要:
【目的】建立水稻叶片蛋白的多反应监测(MRM)质谱绝对定量方法。【方法】水稻叶片蛋白经含1.0%十二烷基硫酸钠(SDS)的磷酸盐(PBS)缓冲液提取,丙酮沉淀除杂纯化、胰蛋白酶消化,酶解液经液相色谱分离,MRM质谱监测,外标法定量。【结果】向提取缓冲液中加入1.0% SDS可增强水稻叶片中16种靶蛋白和总蛋白质的提取效果;不同有机试剂处理,总蛋白质沉淀量存在显著差异(P<0.05),沉淀能力从强到弱依次为乙腈>丙酮>异丙醇>甲醇>乙醇;对于16种目标蛋白,总体以丙酮沉淀效果最好,其次是异丙醇和乙腈,甲醇和乙醇效果较差。该方法线性范围均达到3个数量级,定量限为0.1~2.5 nmol/L,灌浆期16种水稻叶片蛋白质含量为6.0~2818.1 μg/g,相对标准偏差均小于14%。【结论】SDS可显著提高水稻叶片蛋白提取效果,采用丙酮或乙腈可获得较好的蛋白沉淀效果,但不同蛋白质略有差异。结合MRM质谱监测技术可实现水稻叶片蛋白的绝对定量,方法线性范围宽、敏度高、重复性好。
中图分类号:
陈铭学, 杨欢, 曹赵云, 马有宁, 曹珍珍, 程方民. 基于多反应监测质谱技术的水稻叶片蛋白绝对定量方法[J]. 中国水稻科学, 2020, 34(3): 278-286.
Mingxue CHEN, Huan YANG, Zhaoyun CAO, Youning MA, Zhenzhen CAO, Fangmin CHENG. Absolute Quantification of Proteins in Rice Leaf Based on Multiple Reaction Monitoring Mass Spectrometry[J]. Chinese Journal OF Rice Science, 2020, 34(3): 278-286.
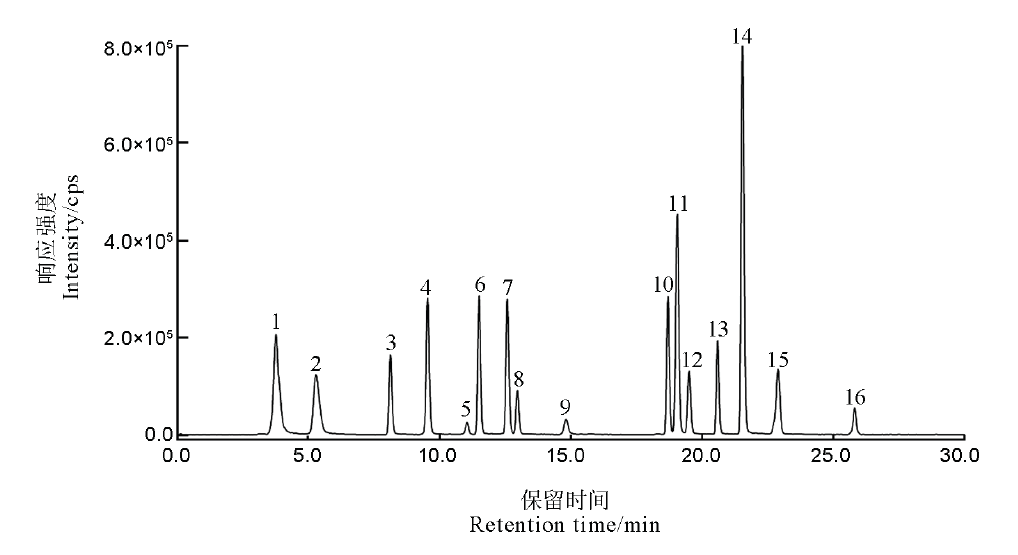
图1 16种特征肽段的总离子流
Fig. 1. Total ion chromatogram of 16 kinds of signature peptides. 1, P12085; 2, Q40677; 3, P0C2Z6; 4, Q8H8D6; 5, Q8GTK4; 6, A3BLC3; 7, Q650W6; 8, Q9ZP20; 9, Q0D5P8; 10, Q948T6; 11, Q8S718; 12, Q7X8A1; 13, Q0DEU8; 14, Q7XV11; 15, Q7XZW5; 16, Q69S39.
| 蛋白质序列号 Accession | 线性方程 Linear equation | 相关系数 r | 线性范围 Linear range/(nmol·L-1) | 检出限 LOD/(nmol·L-1) | 定量限 LOQ/(nmol·L-1) |
|---|---|---|---|---|---|
| Q40677 | y=1.97e4 x + 5.21e5 | 0.9965 | 0.5~250.0 | 0.17 | 0.5 |
| P12085 | y=1.75e4 x + 3.11e5 | 0.9990 | 0.6~300.0 | 0.20 | 0.6 |
| A3BLC3 | y=0.98e4 x + 5.47e5 | 0.9974 | 0.3~200.0 | 0.10 | 0.3 |
| Q9ZP20 | y=3.25e3 x + 2.94e5 | 0.9983 | 1.0~1000.0 | 0.33 | 1.0 |
| Q0D5P8 | y=4.14e3 x + 6.54e5 | 0.9909 | 2.5~1000.0 | 0.80 | 2.5 |
| Q7X8A1 | y=8.59e4 x + 2.95e6 | 0.9993 | 0.5~250.0 | 0.17 | 0.5 |
| Q0DEU8 | y=2.14e3 x + 4.87e5 | 0.9974 | 0.5~250.0 | 0.17 | 0.5 |
| Q7XV11 | y=4.59e3 x + 2.65e5 | 0.9986 | 0.1~100.0 | 0.03 | 0.1 |
| Q7XZW5 | y=2.56e3 x + 2.46e5 | 0.9959 | 0.5~250.0 | 0.17 | 0.5 |
| P0C2Z6 | y=9.13e3 x + 4.48e4 | 0.9967 | 0.7~300.0 | 0.23 | 0.7 |
| Q650W6 | y=7.54e3 x + 6.21e5 | 0.9987 | 0.4~200.0 | 0.13 | 0.4 |
| Q69S39 | y=1.58e3 x + 4.64e5 | 0.9988 | 1.7~1000.0 | 0.56 | 1.7 |
| Q8GTK4 | y=2.51e4 x + 3.36e6 | 0.9989 | 1.8~1000.0 | 0.60 | 1.8 |
| Q8H8D6 | y=5.57e4 x + 3.78e5 | 0.9991 | 0.4~200.0 | 0.13 | 0.4 |
| Q8S718 | y=3.45e4 x + 2.49e5 | 0.9988 | 0.3~200.0 | 0.10 | 0.3 |
| Q948T6 | y=4.44e3 x + 4.25e5 | 0.9982 | 0.4~200.0 | 0.13 | 0.4 |
表1 16种特征肽段的线性方程、相关系数及仪器检出限与定量限
Table 1 Linear equations, correlation coeffcients(r), limits of detection(LOD) and limits of quantification (LOQ) of the 16 kinds of signature peptides.
| 蛋白质序列号 Accession | 线性方程 Linear equation | 相关系数 r | 线性范围 Linear range/(nmol·L-1) | 检出限 LOD/(nmol·L-1) | 定量限 LOQ/(nmol·L-1) |
|---|---|---|---|---|---|
| Q40677 | y=1.97e4 x + 5.21e5 | 0.9965 | 0.5~250.0 | 0.17 | 0.5 |
| P12085 | y=1.75e4 x + 3.11e5 | 0.9990 | 0.6~300.0 | 0.20 | 0.6 |
| A3BLC3 | y=0.98e4 x + 5.47e5 | 0.9974 | 0.3~200.0 | 0.10 | 0.3 |
| Q9ZP20 | y=3.25e3 x + 2.94e5 | 0.9983 | 1.0~1000.0 | 0.33 | 1.0 |
| Q0D5P8 | y=4.14e3 x + 6.54e5 | 0.9909 | 2.5~1000.0 | 0.80 | 2.5 |
| Q7X8A1 | y=8.59e4 x + 2.95e6 | 0.9993 | 0.5~250.0 | 0.17 | 0.5 |
| Q0DEU8 | y=2.14e3 x + 4.87e5 | 0.9974 | 0.5~250.0 | 0.17 | 0.5 |
| Q7XV11 | y=4.59e3 x + 2.65e5 | 0.9986 | 0.1~100.0 | 0.03 | 0.1 |
| Q7XZW5 | y=2.56e3 x + 2.46e5 | 0.9959 | 0.5~250.0 | 0.17 | 0.5 |
| P0C2Z6 | y=9.13e3 x + 4.48e4 | 0.9967 | 0.7~300.0 | 0.23 | 0.7 |
| Q650W6 | y=7.54e3 x + 6.21e5 | 0.9987 | 0.4~200.0 | 0.13 | 0.4 |
| Q69S39 | y=1.58e3 x + 4.64e5 | 0.9988 | 1.7~1000.0 | 0.56 | 1.7 |
| Q8GTK4 | y=2.51e4 x + 3.36e6 | 0.9989 | 1.8~1000.0 | 0.60 | 1.8 |
| Q8H8D6 | y=5.57e4 x + 3.78e5 | 0.9991 | 0.4~200.0 | 0.13 | 0.4 |
| Q8S718 | y=3.45e4 x + 2.49e5 | 0.9988 | 0.3~200.0 | 0.10 | 0.3 |
| Q948T6 | y=4.44e3 x + 4.25e5 | 0.9982 | 0.4~200.0 | 0.13 | 0.4 |
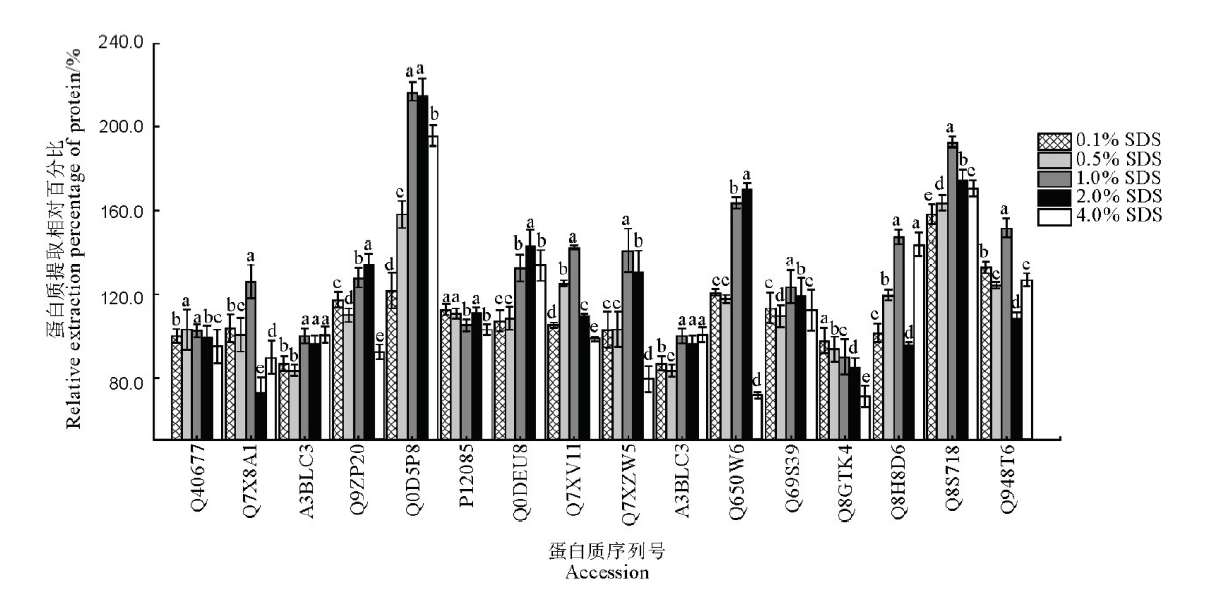
图2 提取缓冲液中不同浓度SDS对16种水稻叶片蛋白提取效果影响图中数据为均值±标准差(n=3);不同字母表示在0.05水平上有显著性差异,而具相同字母表示无显著性差异。纵坐标为蛋白质相对提取百分比,以未加SDS的提取液对应的各目标蛋白提取量为100%计算。
Fig. 2. Influence of different concentrations of SDS in extraction buffer on the extraction effects of 16 kinds of proteins in rice leaves. The data in the figure are mean ± SD (n=3), and different letters indicate significant differences at P<0.05, while the same letters mean no significant difference. The ordinate is the relative extraction percentage of protein with the SDS-free extraction amount of each target protein as 100%.
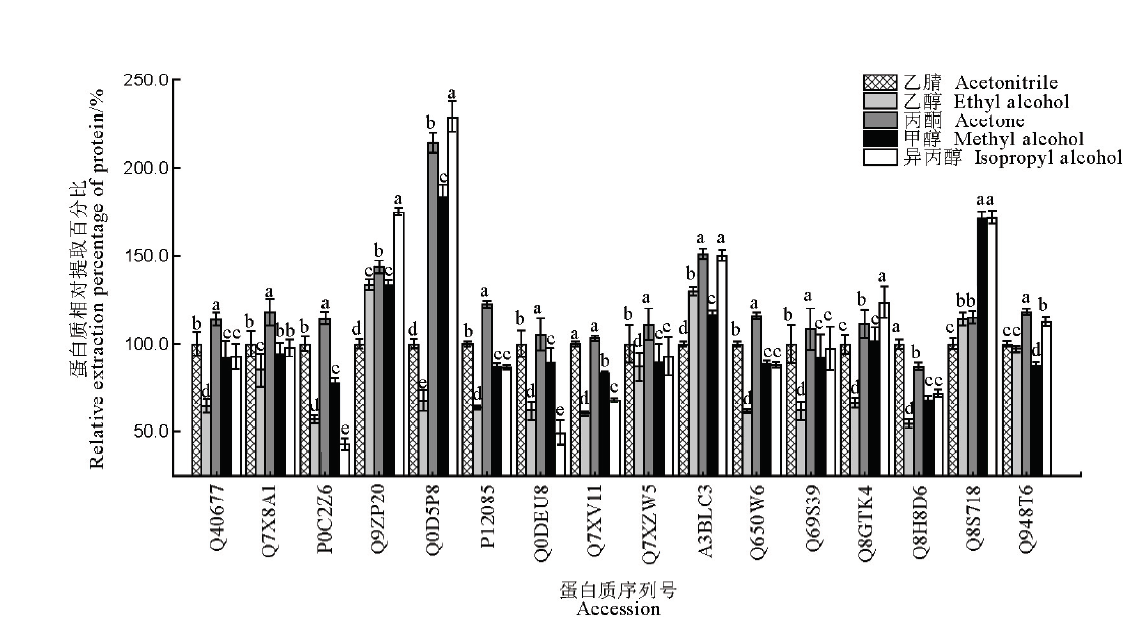
图3 五种有机试剂对16种水稻叶片蛋白沉淀效果比较图中数据为均值±标准差(n=3),不同字母表示在0.05水平上有显著性差异,而具相同字母表示无显著性差异。纵坐标为蛋白质相对提取百分比,以乙腈沉淀对应的各目标蛋白提取量为100%计。
Fig. 3. Comparison of precipitation effects of 16 kinds of proteins in rice leaves among five different organic reagents. The data in the figure are mean ± SD (n=3), and different letters indicate significant differences at P<0.05, while the same letters mean no significant difference. The ordinate is the relative extraction percentage of target protein using various precipitation reagents, with acetonitrile-precipitated protein amount as 100%.
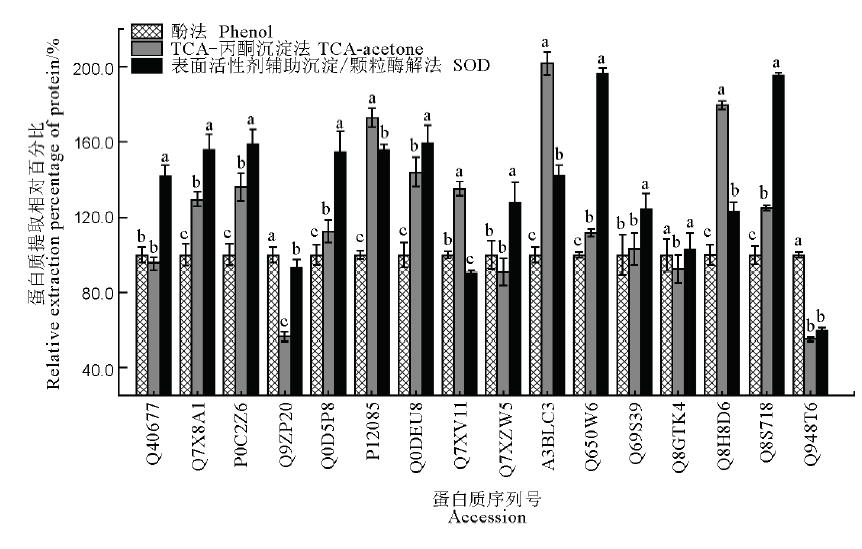
图4 SOD法、酚法和TCA-丙酮沉淀法对16种水稻叶片蛋白的提取效果比较图中数据为均值±标准差(n=3),不同字母表示在0.05水平上有显著性差异,而具相同字母表示无显著性差异。纵坐标为蛋白质相对提取量,以酚法对应的各目标蛋白提取量为100%计。
Fig. 4. Comparison of extraction effects of 16 proteins in rice leaves by SOD, phenol and TCA-acetone precipitation methods. The data in the figure are as mean ± SD (n=3), and different letters indicate significant differences at P<0.05, while the same letters mean no significant difference. The ordinate is the relative extraction percentage of target protein by various methods, with protein amount extracted by phenol method as 100%.
| 蛋白序列号Accession | 测定含量 Measured content/(μg·g-1) | 相对标准偏差 RSD/% |
|---|---|---|
| Q40677 | 589.5±57.5 | 9.8 |
| P12085 | 28.5±2.3 | 8.2 |
| A3BLC3 | 43.3±3.9 | 8.9 |
| Q9ZP20 | 50.2±3.6 | 7.1 |
| Q0D5P8 | 606.9±56.0 | 9.2 |
| Q7X8A1 | 328.3±27.1 | 8.3 |
| Q0DEU8 | 285.8±30.1 | 10.5 |
| Q7XV11 | 20.6±1.7 | 8.2 |
| Q7XZW5 | 153.2±14.0 | 9.1 |
| P0C2Z6 | 67.9±4.8 | 7.0 |
| Q650W6 | 25.5±2.9 | 11.3 |
| Q69S39 | 2818.1±219.9 | 7.8 |
| Q8GTK4 | 109.6±8.1 | 7.4 |
| Q8H8D6 | 6.0±0.4 | 6.2 |
| Q8S718 | 11.7±0.9 | 7.4 |
| Q948T6 | 11.8±1.7 | 13.9 |
表2 SOD法测定水稻16种叶片蛋白质绝对含量(n=6)
Table 2 Determination of absolute contents of 16 kinds of proteins in rice leaves by SOD method(n=6).
| 蛋白序列号Accession | 测定含量 Measured content/(μg·g-1) | 相对标准偏差 RSD/% |
|---|---|---|
| Q40677 | 589.5±57.5 | 9.8 |
| P12085 | 28.5±2.3 | 8.2 |
| A3BLC3 | 43.3±3.9 | 8.9 |
| Q9ZP20 | 50.2±3.6 | 7.1 |
| Q0D5P8 | 606.9±56.0 | 9.2 |
| Q7X8A1 | 328.3±27.1 | 8.3 |
| Q0DEU8 | 285.8±30.1 | 10.5 |
| Q7XV11 | 20.6±1.7 | 8.2 |
| Q7XZW5 | 153.2±14.0 | 9.1 |
| P0C2Z6 | 67.9±4.8 | 7.0 |
| Q650W6 | 25.5±2.9 | 11.3 |
| Q69S39 | 2818.1±219.9 | 7.8 |
| Q8GTK4 | 109.6±8.1 | 7.4 |
| Q8H8D6 | 6.0±0.4 | 6.2 |
| Q8S718 | 11.7±0.9 | 7.4 |
| Q948T6 | 11.8±1.7 | 13.9 |
| [1] | 钱小红. 定量蛋白质组学分析方法[J]. 色谱, 2013, 31(8): 719-723. |
| Qian X H.Quantitative proteomics analysis[J]. Chinese Journal of Chromatography, 2013, 31(8): 719-723. (in Chinese with English abstract) | |
| [2] | 齐林玉, 姜淼, 郭艳, 王迎, 陈平. 质谱MRM技术结合稳定同位素标记法的应用进展[J]. 生命科学研究, 2015, 19(5): 444-451. |
| Qi L Y, Jiang M, Guo Y, Wang Y, Chen P.Progresses on application of MS MRM technology combining with stable isotope labeling[J]. Life Science Research, 2015, 19(5): 444-451. (in Chinese with English abstract) | |
| [3] | Veronika V, Zdenek S.A review on mass spectrometry-based quantitative proteomics: Targeted and data independent acquisition[J]. Analytica Chimica Acta, 2017, 964: 7-23. |
| [4] | 王素兰, 高华萍, 张菁, 叶翔. 基于稳定同位素标记和平行反应监测的蛋白质组学定量技术用于肝癌生物标志物的筛选和验证[J]. 色谱, 2017, 35(9): 934-940. |
| Wang S L, Gao H P, Zhang J, Ye X.Stable isotope labeling and parallel reaction monitoring-based proteomic quantification for biomarker screening and validation of hepatocellular carcinoma[J]. Chinese Journal of Chromatography, 2017, 35(9): 934-940. (in Chinese with English abstract) | |
| [5] | Chen Y T, Chen H W, Wu C F, Chu L J, Chiang W F, Wu C C, Yu J S, Tsai C H, Liang K H, Chang Y S, Wu M, Ou Yang W T. Development of a multiplexed liquid chromatography multiple-reaction-monitoring mass spectrometry (LC-MRM/MS) method for evaluation of salivary proteins as oral cancer biomarkers[J]. Molecular Cellular Proteomics, 2017, 16(5): 799-811. |
| [6] | Zhang J, Lai S, Cai Z, Chen Q, Huang B, Ren Y.Determination of bovine lactoferrin in dairy products by ultra-high performance liquid chromatography-tandem mass spectrometry based on tryptic signature peptides employing an isotope-labeled winged peptide as internal standard[J]. Analytica Chimica Acta, 2014, 4(829): 33-39. |
| [7] | Planque M, Arnould T, Delahaut P, Renardb P, Dieub M, Gillard N.Development of a strategy for the quantification of food allergens in several food products by mass spectrometry in a routine laboratory[J]. Food Chemistry, 2019, 274: 35-45. |
| [8] | Rautengarten C, Ebert B, Heazlewood J L.Absolute quantitation of in vitro expressed plant membrane proteins by targeted proteomics (MRM) for the determination of kinetic parameters[J]. Methods in Molecular Biology, 2018, 1696: 217-234. |
| [9] | Gong C, Zheng N, Zeng J, Aubry A F, Arnold M E.Post-pellet-digestion precipitation and solid phase extraction: A practical and efficient workflow to extract surrogate peptides for ultra-high performance liquid chromatography-tandem mass spectrometry bioanalysis of a therapeutic antibody in the low ng/mL range[J]. Journal of Chromatography A, 2015, 1424: 27-36. |
| [10] | Wu X, Xiong E, Wang W, Scali M, Cresti M.Universal sample preparation method integrating trichloroacetic acid/acetone precipitation with phenol extraction for crop proteomic analysis[J]. Nature Protocols, 2014, 9(2): 362-374. |
| [11] | Luís I M, Alexandre B M, Oliveira M M, Abreu I A.Selection of an appropriate protein extraction method to study the phosphoproteome of maize photosynthetic tissue[J/OL].PLoS One, 2016, 11(10): e0164387. |
| [12] | Kilambi H V, Manda K, Sanivarapu H, Maurya V K, Sharma R, Sreelakshmi Y.Shotgun proteomics of tomato fruits: Evaluation, optimization and validation of sample preparation methods and mass spectrometric parameters [J/OL]. Frontiers in Plant Science, 2016, 7: 969. |
| [13] | Bereman M S, Egertson J D, MacCoss M J. Comparison between procedures using SDS for shotgun proteomic analyses of complex samples[J]. Proteomics, 2011, 11(14): 2931-5. |
| [14] | Yang L, Ji J, Harris-Shultz K R, Wang H, Wang H, Abd-Allah E F, Luo Y, Hu X Y. The dynamic changes of the plasma membrane proteins and the protective roles of nitric oxide in rice subjected to heavy metal cadmium stress[J/OL].Frontiers in Plant Science, 2016, 7: 190. |
| [15] | 高欢欢, 马有宁, 林晓燕, 柴爽爽, 秦美玲, 张涵彤, 何巧, 陈铭学. 亲水作用-反相二维液相色谱串联质谱法鉴定水稻蛋白质[J]. 分析化学, 2018, 46(5): 650-657. |
| Gao H H, Ma Y N, Lin X Y, Chai S S, Qin M L, Zhang H T, He Q, ChenM X. Development of two-dimensional liquid chromatography coupled with tandem mass spectrometry for identification of extracted proteins of rice leaves: Hydrophilic interaction-reversed-phase approach[J]. Chinese Journal of Analytical Chemistry, 2018, 46(5): 650-657. (in Chinese with English abstract) | |
| [16] | Ippoushi K, Sasanuma M, Oike H, Kobori M, Maeda-Yamamoto M.Absolute quantification of protein NP24 in tomato fruit by liquid chromatography/tandem mass spectrometry using stable isotope-labelled tryptic peptide standard[J]. Food Chemistry, 2015, 173: 238-242. |
| [17] | Wisniewski J R, Zouqman A, Nagaraj N, Mann M.Universal sample preparation method for proteome analysis[J]. Nature Methods, 2009, 6(5): 359-362. |
| [18] | Bru-Martínez R, Martínez-Márquez A, Morante-Carriel J, Sellés-Marchart S, Martínez-Esteso M J, Pineda-Lucas J L, Luque I. Targeted quantification of isoforms of a thylakoid-bound protein: MRM method development[J]. Methods in Molecular Biology, 2018, 1696: 147-162. |
| [19] | An B, Zhang M, Johnson R W, Qu J.Surfactant-aided precipitation/on-pellet-digestion (SOD) procedure provides robust and rapid sample preparation for reproducible, accurate and sensitive LC/MS quantification of therapeutic protein in plasma and tissues[J]. Analytical Chemistry, 2015, 87(7): 4023-4029. |
| [20] | Santa C, Anjo S I, Manadas B.Protein precipitation of diluted samples in SDS-containing buffer with acetone leads to higher protein recovery and reproducibility in comparison with TCA/acetone approach[J]. Proteomics, 2016, 16(13): 1847-1851. |
| [21] | Botelho D, Wall M J, Vieira D B, Fitzsimmons S, Liu F, Doucette A.Top-down and bottom-up proteomics of SDS containing solutions following mass-based separation[J]. Journal of Proteome Research, 2010, 9(6): 2863-2870. |
| [22] | Zhang N, Chen R, Young N, Wishart D, Winter P, Weiner J H, Li L.Comparison of SDS and methanol-assisted protein solubilization and digestion methods for Escherichia coli membrane proteome analysis by 2D LC-MS/MS[J]. Proteomics, 2007, 7(4): 484-493. |
| [23] | Ilavenil S, Al-Dhabi N A, Srigopalram S, Kim Y O, Agastian P, BaaruKi R, Choon Choi K C, Arasu M V, Park C G, Park K H. Removal of SDS from biological protein digests for proteomic analysis by mass spectrometry[J]. Proteome Science, 2016, 14(1): 11. |
| [24] | Ouyang Z, Furlong M T, Wu S, Sleczka B, Tamura J, Wang H, Suchard S, Suri A, Olah T, Tymiak A, Jemal M.Pellet digestion: A simple and efficient sample preparation technique for LC-MS/MS quantification of large therapeutic proteinsin plasma[J]. Bioanalysis, 2012, 4(1): 17-28. |
| [25] | Srivastava A K. A Comparison of protein extraction methods using organic solvents for secretome of aspergillus fumigatus strain (MTCC 1811). International Journal of Scientific Research, 2015, 4(6): 2277-8179. |
| [26] | 张冬梅, 马慧萍, 贾正平. 纳升级反相液相色谱串联质谱法分析海马蛋白质组[J]. 药物分析杂志, 2018, 38(1): 118-123. |
| Zhang D M, Ma H P, Jia Z P.Proteomic analysis of Hippocampus using nanoflow reversed phase liquid chromatography-tandem mass spectrometry[J]. Chinese Journal of Pharmaceutical Analysis, 2018, 38(1): 118-123. (in Chinese with English abstract) | |
| [27] | Rezeli M, Sjödin K, Lindberg H, Gidlöf O, Lindahl B, Jernberg T, Spaak J, Erlinge D, Marko-Varga G.Quantitation of 87 proteins by nLC-MRM/MS in human plasma: workflow for large-scale analysis of biobank samples[J]. Journal of Proteome Research, 2017, 16(19): 3242-3254. |
| [28] | Hoofnagle A N, Becker J O, Oda M N, Cavigiolio G, Mayer P, Vaisar T.Multiple-reaction monitoring-mass spectrometric assays can accurately measure the relative protein abundance in complex mixture[J]. Clinical Chemistry, 2012, 58(4): 777-781. |
| [1] | 陆丹丹, 雍明玲, 陶钰, 叶苗, 张祖建. 优良食味水稻品种籽粒蛋白质积累特征及其对氮素水平的响应[J]. 中国水稻科学, 2022, 36(5): 520-530. |
| [2] | 王志东, 陈宜波, 龚蓉, 周少川, 王重荣, 李宏, 黄道强, 周德贵, 赵雷, 潘阳阳, 杨义强, 李晓芳. 优质籼稻剑叶SPAD值与稻米品质相关性研究[J]. 中国水稻科学, 2021, 35(1): 89-97. |
| [3] | 石吕, 张新月, 孙惠艳, 曹先梅, 刘建, 张祖建. 不同类型水稻品种稻米蛋白质含量与蒸煮食味品质的关系及后期氮肥的效应[J]. 中国水稻科学, 2019, 33(6): 541-552. |
| [4] | 何婧, 刘寒, 郭留明, 李静, 张恒木. 水稻小热休克蛋白OsHSP20抗体特性鉴定及应用[J]. 中国水稻科学, 2019, 33(3): 235-240. |
| [5] | 张敏娟, 李帅军, 陈琼琼, 景秀清, 陈坤明, 石春海, 李文强. 水稻矮化少蘖突变体dlt3的基因定位和蛋白质组学分析[J]. 中国水稻科学, 2018, 32(6): 529-537. |
| [6] | 黄小平, 张宏玉, 雷刚, 王志美, 章智, 贺超, 廖江林, 黄英金. 灌浆期夜间高温胁迫下耐热和热敏感水稻籽粒的比较蛋白质组分析[J]. 中国水稻科学, 2017, 31(1): 13-22. |
| [7] | 刘立军1,*,王康君2,卞金龙1,熊溢伟1,王志琴1,杨建昌1. 结实期干湿交替灌溉对籽粒蛋白质含量不同的转基因水稻的生理特性及产量的影响[J]. 中国水稻科学, 2014, 28(4): 384-390. |
| [8] | 梁成刚#,张青#,李敬,熊丹,许光利,汪燕,刘泉,黄鹏,李天*. 水稻灌浆期高温对天冬氨酸代谢酶活性及其家族氨基酸含量的影响[J]. 中国水稻科学, 2013, 27(1): 71-76. |
| [9] | 谢黎虹#,罗炬#,唐绍清,陈能,焦桂爱,邵高能 ,魏祥进,胡培松*. 蛋白质影响水稻米饭食味品质的机理[J]. 中国水稻科学, 2013, 27(1): 91-96. |
| [10] | 文李1, 3,*,刘盖2,王坤2 ,谭碧君1 ,陶钧1. 红莲型细胞质雄性不育水稻单核期花粉蛋白差异表达分析[J]. 中国水稻科学, 2012, 26(5): 529-536. |
| [11] | 邵彩虹,钱银飞,唐秀英,刘光荣 ,谢金水,彭春瑞,邱才飞. 养分胁迫对水稻籽粒灌浆充实影响的蛋白质组学研究[J]. 中国水稻科学, 2012, 26(3): 267-274. |
| [12] | 杨亚春,倪大虎,宋丰顺,李莉,冯光,李泽福,杨剑波*. 两个环境下糙米和精米蛋白质含量的QTL分析[J]. 中国水稻科学, 2012, 26(3): 351-355. |
| [13] | 彭春瑞1,2,#邵彩虹2,# 潘晓华1,* 钱银飞2邱才飞2谢金水2. 水稻育秧肥的壮秧效应及其蛋白质组学分析[J]. 中国水稻科学, 2012, 26(1): 27-33. |
| [14] | 董明辉,谢裕林,乔中英 ,刘晓斌,吴翔宙,赵步洪,杨建昌,. 水稻不同粒位籽粒淀粉与蛋白质累积动态差异[J]. 中国水稻科学, 2011, 25(3): 297-306 . |
| [15] | 谢金水, 邵彩虹, 唐秀英, 石庆华,. 养分胁迫对籽粒灌浆期水稻叶片衰老影响的蛋白质组学分析[J]. 中国水稻科学, 2011, 25(2): 143-149 . |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||