
中国水稻科学 ›› 2019, Vol. 33 ›› Issue (2): 144-151.DOI: 10.16819/j.1001-7216.2019.8120
朱安东1, 孙志超1, 朱玉君1, 张荟2,3, 牛小军1, 樊叶杨1, 张振华1, 庄杰云1,*( )
)
收稿日期:2018-11-06
修回日期:2018-12-14
出版日期:2019-03-10
发布日期:2019-03-10
通讯作者:
庄杰云
基金资助:
Andong ZHU1, Zhichao SUN1, Yujun ZHU1, Hui ZHANG2,3, Xiaojun NIU1, Yeyang FAN1, Zhenhua ZHANG1, Jieyun ZHUANG1,*( )
)
Received:2018-11-06
Revised:2018-12-14
Online:2019-03-10
Published:2019-03-10
Contact:
Jieyun ZHUANG
摘要:
【目的】粒重粒形是影响水稻产量和品质的重要因素,由大量数量性状座位(QTL)控制,其作用变异极大,但以往研究主要着眼于效应大的QTL。本研究在剔除主效QTL影响的基础上,开展微效粒重粒形QTL分析。【方法】在前期研究基础上,从原群体挑选出1个剩余杂合体单株,构建了在主效QTL区间纯合、在其余区域中13个区间分离的群体,种植于浙江杭州和海南陵水,测定千粒重、粒长和粒宽。【结果】采用Windows QTL Cartographer 2.5,检测到22个QTL,分布于10条染色体的12个区间,其中,10个区间在两地均呈显著作用,2个区间仅在杭州试验中呈显著作用。进一步从该群体筛选出1个只在其中4个QTL区间杂合的单株,自交构建分离群体,验证了这4个区间对粒重粒形的效应。【结论】排除主效QTL有利于提高微效粒重粒形QTL的检测功效;虽然微效QTL可能易受环境和遗传背景影响,但仍可具有稳定表现。这些结果为进一步开展粒重粒形QTL的精细定位、克隆和分子标记辅助选择奠定了基础。
中图分类号:
朱安东, 孙志超, 朱玉君, 张荟, 牛小军, 樊叶杨, 张振华, 庄杰云. 应用剩余杂合体衍生群体定位水稻粒重粒形QTL[J]. 中国水稻科学, 2019, 33(2): 144-151.
Andong ZHU, Zhichao SUN, Yujun ZHU, Hui ZHANG, Xiaojun NIU, Yeyang FAN, Zhenhua ZHANG, Jieyun ZHUANG. Identification of QTL for Grain Weight and Grain Shape Using Populations Derived from Residual Heterozygous Lines of indica Rice[J]. Chinese Journal OF Rice Science, 2019, 33(2): 144-151.
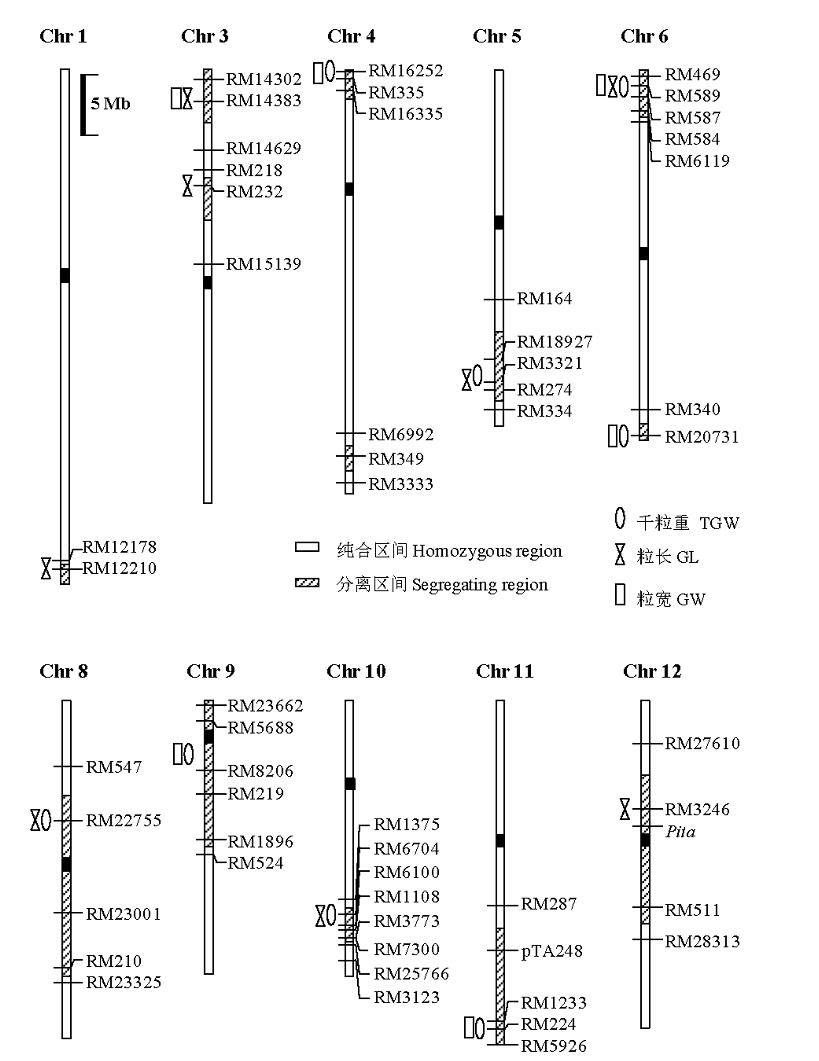
图1 在Ti52-2群体检测到的千粒重、粒长和粒宽QTL在染色体上的位置
Fig. 1. Chromosomal regions of QTL for the 1000-grain weight (TGW), grain length (GL) and grain width (GW) detected in the Ti52-2 population.
| 性状 Trait | 群体 Population | 地点 Location | 株系数 No. of lines | 平均值 Mean | 标准差 SD | 变异系数 CV | 变异范围 Range | 偏度 Skewness | 峰度 Kurtosis | 亲本Parent | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 特青Teqing | IRBB52 | ||||||||||
| 千粒重 | Ti52-2 | 陵水 Lingshui | 251 | 24.98 | 0.51 | 0.020 | 23.77-26.32 | -0.03 | -0.21 | 25.53 | 24.87 |
| TGW / g | Ti52-2 | 杭州 Hangzhou | 251 | 21.83 | 0.42 | 0.019 | 20.78-23.00 | 0.25 | -0.46 | 23.55 | 23.14 |
| ZC8 | 杭州 Hangzhou | 179 | 22.18 | 0.27 | 0.012 | 21.46-22.93 | 0.29 | 0.09 | 22.95 | 23.52 | |
| 粒长 | Ti52-2 | 陵水 Lingshui | 251 | 8.009 | 0.104 | 0.013 | 7.742-8.382 | 0.21 | 0.21 | 7.541 | 9.026 |
| GL / mm | Ti52-2 | 杭州 Hangzhou | 251 | 7.821 | 0.094 | 0.012 | 7.567-8.053 | 0.14 | -0.29 | 7.417 | 8.911 |
| ZC8 | 杭州 Hangzhou | 179 | 8.137 | 0.053 | 0.007 | 8.016-8.342 | 0.22 | 0.36 | 7.650 | 9.122 | |
| 粒宽 | Ti52-2 | 陵水 Lingshui | 251 | 2.890 | 0.039 | 0.013 | 2.777-2.997 | -0.11 | 0.00 | 3.216 | 2.720 |
| GW / mm | Ti52-2 | 杭州 Hangzhou | 251 | 2.579 | 0.028 | 0.011 | 2.491-2.652 | 0.05 | -0.09 | 2.891 | 2.395 |
| ZC8 | 杭州 Hangzhou | 179 | 2.688 | 0.016 | 0.006 | 2.646-2.728 | 0.20 | -0.34 | 2.917 | 2.510 | |
表1 Ti52-2群体的千粒重、粒长和粒宽表现
Table 1 Phenotypic performance of 1000-grain weight(TGW), grain length(GL) and grain width(GW) in Ti52-2 population.
| 性状 Trait | 群体 Population | 地点 Location | 株系数 No. of lines | 平均值 Mean | 标准差 SD | 变异系数 CV | 变异范围 Range | 偏度 Skewness | 峰度 Kurtosis | 亲本Parent | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 特青Teqing | IRBB52 | ||||||||||
| 千粒重 | Ti52-2 | 陵水 Lingshui | 251 | 24.98 | 0.51 | 0.020 | 23.77-26.32 | -0.03 | -0.21 | 25.53 | 24.87 |
| TGW / g | Ti52-2 | 杭州 Hangzhou | 251 | 21.83 | 0.42 | 0.019 | 20.78-23.00 | 0.25 | -0.46 | 23.55 | 23.14 |
| ZC8 | 杭州 Hangzhou | 179 | 22.18 | 0.27 | 0.012 | 21.46-22.93 | 0.29 | 0.09 | 22.95 | 23.52 | |
| 粒长 | Ti52-2 | 陵水 Lingshui | 251 | 8.009 | 0.104 | 0.013 | 7.742-8.382 | 0.21 | 0.21 | 7.541 | 9.026 |
| GL / mm | Ti52-2 | 杭州 Hangzhou | 251 | 7.821 | 0.094 | 0.012 | 7.567-8.053 | 0.14 | -0.29 | 7.417 | 8.911 |
| ZC8 | 杭州 Hangzhou | 179 | 8.137 | 0.053 | 0.007 | 8.016-8.342 | 0.22 | 0.36 | 7.650 | 9.122 | |
| 粒宽 | Ti52-2 | 陵水 Lingshui | 251 | 2.890 | 0.039 | 0.013 | 2.777-2.997 | -0.11 | 0.00 | 3.216 | 2.720 |
| GW / mm | Ti52-2 | 杭州 Hangzhou | 251 | 2.579 | 0.028 | 0.011 | 2.491-2.652 | 0.05 | -0.09 | 2.891 | 2.395 |
| ZC8 | 杭州 Hangzhou | 179 | 2.688 | 0.016 | 0.006 | 2.646-2.728 | 0.20 | -0.34 | 2.917 | 2.510 | |
| 染色体 Chr | 区间 Interval | QTL | 杭州 Hangzhou | 陵水 Lingshui | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LOD | A | D | R2/% | LOD | A | D | R2/% | |||
| 1 | RM12210 | qGL1 | 5.69 | 0.029 | -0.004 | 4.51 | 4.55 | 0.033 | 0.004 | 4.76 |
| 3 | RM14302-RM14383 | qGL3.1 | 4.90 | 0.025 | 0.003 | 3.52 | 8.12 | 0.044 | 0.013 | 9.22 |
| qGW3 | ns | 5.29 | -0.015 | 0.001 | 6.93 | |||||
| 3 | RM232 | qGL3.2 | 5.21 | 0.029 | -0.004 | 3.79 | ns | |||
| 4 | RM16252-RM335 | qTGW4 | 6.65 | -0.150 | 0.090 | 6.66 | 5.03 | -0.200 | -0.030 | 7.14 |
| qGW4 | 9.78 | -0.014 | 0.003 | 11.50 | 6.79 | -0.017 | 0.001 | 9.02 | ||
| 5 | RM18927-RM3321 | qTGW5 | 22.38 | 0.340 | -0.040 | 27.44 | ns | |||
| qGL5 | 38.86 | 0.092 | -0.022 | 40.16 | 6.84 | 0.044 | 0.004 | 7.72 | ||
| 6 | RM469-RM587 | qTGW6.1 | ns | 2.91 | 0.130 | 0.110 | 3.98 | |||
| qGL6 | 13.27 | 0.047 | 0.001 | 11.20 | 10.50 | 0.055 | 0.001 | 11.93 | ||
| qGW6.1 | 12.63 | -0.016 | 0.006 | 14.78 | ns | |||||
| 6 | RM20731 | qTGW6.2 | 4.64 | -0.140 | 0.040 | 4.62 | ns | |||
| qGW6.2 | 5.65 | -0.011 | 0.002 | 6.31 | 5.78 | -0.015 | -0.006 | 7.31 | ||
| 8 | RM22755-RM210 | qTGW8 | 3.84 | 0.120 | 0.020 | 3.55 | 4.49 | 0.170 | -0.090 | 6.29 |
| qGL8 | 5.45 | 0.028 | 0.005 | 4.09 | 5.15 | 0.027 | -0.030 | 5.70 | ||
| 9 | RM5688-RM219 | qTGW9 | 12.30 | 0.230 | 0.050 | 14.23 | ns | |||
| qGW9 | 10.54 | 0.014 | 0.004 | 13.16 | ns | |||||
| 10 | RM6704-RM6100 | qTGW10 | 6.54 | 0.160 | -0.050 | 6.44 | ns | |||
| qGL10 | 9.17 | 0.037 | -0.001 | 7.47 | 6.43 | 0.030 | 0.029 | 6.24 | ||
| 11 | RM1233-RM5926 | qTGW11 | ns | 4.08 | -0.170 | 0.090 | 6.13 | |||
| qGW11 | 4.41 | -0.008 | -0.005 | 4.79 | 7.46 | -0.020 | 0.003 | 10.09 | ||
| 12 | RM3246-Pita | qGL12 | 3.33 | -0.021 | -0.002 | 2.31 | 10.36 | -0.051 | -0.010 | 11.67 |
表2 在Ti52-2群体检测到的千粒重、粒长和粒宽QTL
Table 2 QTL for 1000-grain weight, grain length and grain width detected in the Ti52-2 population.
| 染色体 Chr | 区间 Interval | QTL | 杭州 Hangzhou | 陵水 Lingshui | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LOD | A | D | R2/% | LOD | A | D | R2/% | |||
| 1 | RM12210 | qGL1 | 5.69 | 0.029 | -0.004 | 4.51 | 4.55 | 0.033 | 0.004 | 4.76 |
| 3 | RM14302-RM14383 | qGL3.1 | 4.90 | 0.025 | 0.003 | 3.52 | 8.12 | 0.044 | 0.013 | 9.22 |
| qGW3 | ns | 5.29 | -0.015 | 0.001 | 6.93 | |||||
| 3 | RM232 | qGL3.2 | 5.21 | 0.029 | -0.004 | 3.79 | ns | |||
| 4 | RM16252-RM335 | qTGW4 | 6.65 | -0.150 | 0.090 | 6.66 | 5.03 | -0.200 | -0.030 | 7.14 |
| qGW4 | 9.78 | -0.014 | 0.003 | 11.50 | 6.79 | -0.017 | 0.001 | 9.02 | ||
| 5 | RM18927-RM3321 | qTGW5 | 22.38 | 0.340 | -0.040 | 27.44 | ns | |||
| qGL5 | 38.86 | 0.092 | -0.022 | 40.16 | 6.84 | 0.044 | 0.004 | 7.72 | ||
| 6 | RM469-RM587 | qTGW6.1 | ns | 2.91 | 0.130 | 0.110 | 3.98 | |||
| qGL6 | 13.27 | 0.047 | 0.001 | 11.20 | 10.50 | 0.055 | 0.001 | 11.93 | ||
| qGW6.1 | 12.63 | -0.016 | 0.006 | 14.78 | ns | |||||
| 6 | RM20731 | qTGW6.2 | 4.64 | -0.140 | 0.040 | 4.62 | ns | |||
| qGW6.2 | 5.65 | -0.011 | 0.002 | 6.31 | 5.78 | -0.015 | -0.006 | 7.31 | ||
| 8 | RM22755-RM210 | qTGW8 | 3.84 | 0.120 | 0.020 | 3.55 | 4.49 | 0.170 | -0.090 | 6.29 |
| qGL8 | 5.45 | 0.028 | 0.005 | 4.09 | 5.15 | 0.027 | -0.030 | 5.70 | ||
| 9 | RM5688-RM219 | qTGW9 | 12.30 | 0.230 | 0.050 | 14.23 | ns | |||
| qGW9 | 10.54 | 0.014 | 0.004 | 13.16 | ns | |||||
| 10 | RM6704-RM6100 | qTGW10 | 6.54 | 0.160 | -0.050 | 6.44 | ns | |||
| qGL10 | 9.17 | 0.037 | -0.001 | 7.47 | 6.43 | 0.030 | 0.029 | 6.24 | ||
| 11 | RM1233-RM5926 | qTGW11 | ns | 4.08 | -0.170 | 0.090 | 6.13 | |||
| qGW11 | 4.41 | -0.008 | -0.005 | 4.79 | 7.46 | -0.020 | 0.003 | 10.09 | ||
| 12 | RM3246-Pita | qGL12 | 3.33 | -0.021 | -0.002 | 2.31 | 10.36 | -0.051 | -0.010 | 11.67 |
| 染色体Chr | 区间 Interval | QTL | LOD | A | D | R2/% |
|---|---|---|---|---|---|---|
| 6 | RM20731 | qTGW6.2 | 2.52 | -0.080 | 0.010 | 5.08 |
| qGW6.2 | 4.31 | -0.006 | -0.000 | 8.03 | ||
| 8 | RM22755-RM210 | qTGW8 | 6.52 | 0.160 | -0.010 | 16.32 |
| qGL8 | 5.71 | 0.026 | -0.013 | 11.92 | ||
| 10 | RM1108-RM7300 | qGL10 | 4.70 | 0.024 | 0.000 | 9.27 |
| 12 | RM3246-Pita | qTGW12 | 2.75 | 0.080 | 0.050 | 5.49 |
| qGL12 | 3.58 | -0.019 | 0.008 | 6.79 | ||
| qGW12 | 3.70 | 0.006 | 0.001 | 6.85 |
表3 在ZC8群体检测到的千粒重、粒长和粒宽QTL
Table 3 QTL for 1000-grain weight, grain length and grain width detected in the ZC8 population.
| 染色体Chr | 区间 Interval | QTL | LOD | A | D | R2/% |
|---|---|---|---|---|---|---|
| 6 | RM20731 | qTGW6.2 | 2.52 | -0.080 | 0.010 | 5.08 |
| qGW6.2 | 4.31 | -0.006 | -0.000 | 8.03 | ||
| 8 | RM22755-RM210 | qTGW8 | 6.52 | 0.160 | -0.010 | 16.32 |
| qGL8 | 5.71 | 0.026 | -0.013 | 11.92 | ||
| 10 | RM1108-RM7300 | qGL10 | 4.70 | 0.024 | 0.000 | 9.27 |
| 12 | RM3246-Pita | qTGW12 | 2.75 | 0.080 | 0.050 | 5.49 |
| qGL12 | 3.58 | -0.019 | 0.008 | 6.79 | ||
| qGW12 | 3.70 | 0.006 | 0.001 | 6.85 |
| [1] | 李一博, 赵雷. 水稻品质性状的遗传改良及其关键科学问题. 生命科学, 2016, 28(10): 1168-1179. |
| Li Y B, Zhao L.Genetic improvement and key scientific questions of grain quality traits in rice.Chin Sci Bull, 2016, 28(10): 1168-1179. (in Chinese with English abstract) | |
| [2] | Li N, Xu R, Li Y.Control of grain size in rice.Plant Reprod, 2018, 31(3): 237-251. |
| [3] | Yu J, Xiong H, Zhu X, Zhang H, Li H, Miao J, Wang W, Tang Z, Zhang Z, Yao G, Zhang Q, Pan Y, Wang X, Rashid M A R, Li J, Gao Y, Li Z, Yang W, Fu X, Li Z,. OsLG3 contributing to rice grain length and yield was mined by Ho-LAMap. BMC Biol, 2017, 15(1): 28. |
| [4] | Hu Z, Lu S J, Wang M J, He H, Sun L, Wang H, Liu X H, Jiang L, Sun J L, Xin X, Kong W, Chu C, Xue H W, Yang J, Luo X, Liu J X.A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol Plant, 2018, 11(5): 736-749. |
| [5] | Yu J, Miao J, Zhang Z, Xiong H, Zhu X, Sun X, Pan Y, Liang Y, Zhang Q, Rashid M A R, Li J, Zhang H, Li Z. Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotechnol J, 2018, 16(9): 1667-1678. |
| [6] | Zhao D S, Li Q F, Zhang C Q, Zhang C, Yang Q Q, Pan L X, Ren X Y, Lu J, Gu M H, & Liu Q Q. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat Commun, 2018, 9(1): 1240. |
| [7] | Li Y, Fan C, Xing Y, Jiang Y, Lou L, Sun L, Shao D, Xu C, Li X, Xiao J, He Y, Zhang Q.Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet, 2011, 43: 1266-1269. |
| [8] | Noriko K, Masayuki K, Kei K, Takuya K, Tsutomu N, Yuji H, Itsuro T, Takashi S, Kiyoaki K.Identification of quantitative trait loci for rice grain quality and yield-related traits in two closely related Oryza sativa L. subsp. japonica cultivars grown near the northernmost limit for rice paddy cultivation. Breed Sci, 2017, 67: 191-206. |
| [9] | Dong Q, Zhang Z H, Wang L L, Zhu Y J, Fan Y Y, Mou T M, Ma L Y, Zhuang J Y.Dissection and fine-mapping of two QTL for grain size linked in a 460-kb region on chromosome 1 of rice.Rice, 2018, 11: 44. |
| [10] | Yamamoto T, Yonemaru J, Yano M.Towards the understanding of complex traits in rice: Substantially or superficially?DNA Res, 2009, 16(3): 141-154. |
| [11] | Takai T, Ikka T, Kondo K, Nonoue Y, Ono N, Arai-Sanoh Y, Yoshinaga S, Nakano H, Yano M, Kondo M, Yamamoto T.Genetic mechanisms underlying yield potential in the rice high-yielding cultivar Takanari, based on reciprocal chromosome segment substitution lines.BMC Plant Biol, 2014, 14(1): 295. |
| [12] | Nagata K, Ando T, Nonoue Y, Mizubayashi T, Kitazawa N, Shomura A, Matsubara K, Ono N, Mizobuchi R, Shabaya T, Ogisotanaka E, Hori K, Yano M, Fukuoka S.Advanced backcross QTL analysis reveals complicated genetic control of rice grain shape in a japonica × indica cross. Breed Sci, 2015, 65(4): 308-318. |
| [13] | Ye H, Foley M E, Gu X Y.New seed dormancy loci detected from weedy rice-derived advanced populations with major QTL alleles removed from the background.Plant Sci, 2010, 179(6): 612-619. |
| [14] | Xu F F, Sun C X, Huang Y, Chen Y L, Tong C, Bao J S.QTL mapping for rice grain quality: A strategy to detect more QTLs within sub-populations.Mol Breed, 2015, 35(4): 105. |
| [15] | Wang Z, Chen J Y, Zhu Y J, Fan Y Y, Zhuang J Y.Validation of qGS10, a quantitative trait locus for grain size on the long arm of chromosome 10 in rice(Oryza sativa L.). J Integr Agric, 2017, 16(1): 16-26. |
| [16] | Sun Z C, Zhu Y J, Chen J Y, Zhang H, Zhang Z H, Niu X J, Fan Y Y, Zhuang J Y.Minor-effect QTL for heading date detected in crosses between indica rice cultivar Teqing and near isogenic lines of IR24.Crop J, 2018, 6(3): 291-298. |
| [17] | Zhang H W, Fan Y Y, Zhu Y J, Chen J Y, Yu S B, Zhuang J Y.Dissection of the qTGW1.1 region into two tightly-linked minor QTLs having stable effects for grain weight in rice. BMC Genet, 2016, 17(1): 98. |
| [18] | Zheng K L, Huang N, Bennett J, Khush G S.PCR-based marker-assisted selection in rice breeding// IRRI Discussion Paper Series No.12. Manila, Los Banos, Philippines: International Rice Research Institute, 1995. |
| [19] | Chen X, Temnykh S, Xu Y, Cho Y G, McCouch S R. Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor Appl Genet, 1997, 95(4): 553-567. |
| [20] | Lander E S, Green P, Abrahamson J, Barlow A, Daly M J, Lincoln S E, Newberg L A.MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations.Genomics, 1987, 1(2): 174-181. |
| [21] | Wang S, Basten C J, Zeng Z B.Windows QTL Cartographer 2.5. Raleigh, NC, USA: Department of Statistics, North Carolina State University, 2012. |
| [22] | McCouch S R, CGSNL. Gene nomenclature system for rice.Rice, 2008, 1(1): 72-84. |
| [23] | Huang N, Parco A, Mew T, Magpantay G, McCouch S, Guiderdoni E, Xu J, Subudhi P, Angeles E R, Khush G S. RFLP mapping of isozymes, RAPD and QTLs for grain shape, brown planthopper resistance in a double haploid rice population.Mol Breed, 1997, 3(2): 105-113. |
| [24] | 邢永忠, 谈移芳, 徐才国, 华金平, 孙新立. 利用水稻重组自交系群体定位谷粒外观性状的数量性状基因. 植物学报, 2001, 43(8): 840-845. |
| Xing Y Z, Tan Y F, Xu C G, Hua J P, Sun X L.Mapping quantitative trait loci for grain appearance traits of rice using a recombinant inbred line population.Acta Bot Sin, 2001, 43(8): 840-845. (in Chinese with English abstract) | |
| [25] | Li J X, Yu S B, Xu C G, Tan Y F, Gao Y J, Li X H, Zhang Q.Analyzing quantitative trait loci for yield using a vegetatively replicated F2 population from a cross between the parents of an elite rice hybrid.Theor Appl Genet, 2000, 101: 248-254. |
| [26] | 姜恭好, 徐才国, 李香花, 何予卿. 利用双单倍体群体剖析水稻产量及其相关性状的遗传基础. 遗传学报, 2004, 31(1): 63-72. |
| Jiang G H, Xu C G, Li X H, He Y Q.Characterization of the genetic basis for yield and its component traits of rice revealed by doubled haploid population.Acta Genet Sin, 2004, 31(1): 63-72. (in Chinese with English abstract) | |
| [27] | 王军, 朱金燕, 周勇, 杨杰, 范方军, 李文奇, 梁国华, 仲维功. 基于染色体单片段代换系的水稻粒形QTL定位. 作物学报, 2013, 39(4): 617-625. |
| Wang J, Zhu J Y, Zhou Y, Yang J, Fan F J, Li W Q, Liang G H, Zhong W G.Mapping of QTLs for grain shape using chromosome single segment substitution lines in rice (Oryza sativa L.). Acta Agron Sin, 2013, 39(4): 617-625. (in Chinese with English abstract) | |
| [28] | 林荔辉, 吴为人. 水稻粒型和粒重的QTL定位分析. 分子植物育种, 2003, 1(3): 337-342. |
| Lin L H, Wu W R.Mapping of QTLs underlying grain shape and grain weight in rice.Mol Plant Breed, 2003, 1(3): 337-342. (in Chinese with English abstract) | |
| [29] | Liang Y S, Zhan X D, Gao Z Q, Lin Z C, Yang Z L, Zhang Y X, Shen X H, Cao L Y, Cheng S H.Mapping of QTLs associated with important agronomic traits using three populations derived from a super hybrid rice Xieyou9308.Euphytica, 2012, 184(1): 1-13. |
| [30] | Marathi B, Guleria S, Mohapatra T, Parsad R, Mariappan N, Kurungara V K, Atwal S S, Prabhu K V, Singh N K, Singh A K.QTL analysis of novel genomic regions associated with yield and yield related traits in new plant type based recombinant inbred lines of rice (Oryza sativa L.). BMC Plant Biol, 2012, 12: 137. |
| [31] | Gao F Y, Zeng L H, Qiu L, Lu X J, Ren J S, Wu X T, Su X W, Gao Y M, Ren G J.QTL mapping of grain appearance quality traits and grain weight using a recombinant inbred population in rice (Oryza sativa L.). J Integr Agric, 2016, 15(8): 1693-1702. |
| [32] | Gao Y, Zhu J, Song Y, He C, Shi C, Xing Y.Analysis of digenic epistatic effects and QE interaction effects QTL controlling grain weight in rice. J Zhejiang Univ Sci, 2004, 5(4): 371-377. |
| [33] | Kang Y J, Shim K C, Lee H S, Jeon Y A, Kim S H, Kang J W, Yun Y T, Park I K, Ahn S N.Fine mapping and candidate gene analysis of the quantitative trait locus gw8.1 associated with grain length in rice. Genes & Genom, 2018, 40(4): 389-397. |
| [34] | Tian F, Li D J, Fu Q, Zhu Z F, Fu Y C, Wang X K, Sun C Q.Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice(Oryza sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor Appl Genet, 2006, 112(3): 570-580. |
| [35] | 张亚东, 张颖慧, 董少玲, 陈涛, 赵庆勇, 朱镇, 周丽慧, 姚姝, 赵凌, 于新, 王才林. 特大粒水稻材料粒型性状QTL检测. 中国水稻科学, 2013, 27(2): 122-128. |
| Zhang Y D, Zhang H Y, Dong S L, Chen T, Zhao Q Y, Zhu Z, Zhou L H, Yao S, Zhao L, Yu X, Wang C L.Identification of QTL for rice grain traits based on extra-large grain material.Chin J Rice Sci, 2013, 27(2): 122-128. (in Chinese with English abstract) | |
| [36] | Hittalmani S, Shashidhar H E, Bagali P G, Huang N, Sidhu J S, Singh V P, Khush G S.Molecular mapping of quantitative trait loci for plant growth, yield and yield related traits across three diverse locations in a doubled haploid rice population. Euphytica, 2002, 125(2): 207-214. |
| [37] | Li S, Cui G, Guan C, Wang J, Lian G.QTL detection for rice grain shape using chromosome single segment substitution lines.Rice Sci, 2011, 18(4): 273-278. |
| [38] | 杨占烈, 戴高兴, 翟荣荣, 林泽川, 王会民, 曹立勇, 程式华. 多环境条件下超级杂交稻协优9308重组自交系群体粒形性状QTL定位. 中国水稻科学, 2013, 27(5): 482-490. |
| Yang Z L, Dai G X, Zhai R R, Lin Z C, Wang H M, Cao L Y, Cheng S H.QTL analysis of rice grain shape traits by using recombinant inbred lines from super hybrid rice Xieyou 9308 in multi-environments.Chin J Rice Sci, 2013, 27(5): 482-490. (in Chinese with English abstract) | |
| [39] | Oh J M, Balkunde S, Yang P, Yoon D B, Ahn S N.Fine mapping of grain weight QTL, tgw11 using near isogenic lines from a cross between Oryza sativa and O. grandiglumis. Genes & Genom, 2011, 33(3): 259-265. |
| [40] | 周梦玉, 宋昕蔚, 徐静, 付雪, 李婷, 朱雨晨, 肖幸运, 毛一剑, 曾大力, 胡江, 朱丽, 任德勇, 高振宇, 郭龙彪, 钱前, 吴明国, 林建荣, 张光恒. 籼稻C84和粳稻春江16B重组自交系遗传图谱构建及籽粒性状QTL定位与验证. 中国水稻科学, 2018, 32(3): 207-218. |
| Zhou M Y, Song X W, Xu J, Fu X, Li T, Zhu Y C, Xiao X Y, Mao Y J, Zeng D L, Hu J, Zhu L, Ren D Y, Gao Z Y, Guo L B, Qian Q, Wu M G, Lin J R, Zhang G H.Construction of genetic map and mapping and verification of grain traits QTLs using recombinant inbred lines derived from a cross between indica C84 and japonica CJ16B. Chin J Rice Sci, 2018, 32(3): 207-218. (in Chinese with English abstract) |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||