
中国水稻科学 ›› 2019, Vol. 33 ›› Issue (1): 1-11.DOI: 10.16819/j.1001-7216.2018.8026
• 研究论文 • 下一篇
林添资1,2, 孙立亭2, 龚红兵2, 王益华1, 刘玲珑1, 赵志刚1, 江玲1, 万建民1,*( )
)
收稿日期:2018-03-13
修回日期:2018-05-29
出版日期:2019-01-10
发布日期:2019-01-10
通讯作者:
万建民
基金资助:
Tianzi LIN1,2, Liting SUN2, Hongbin GONG2, Yihua WANG1, Linglong LIU1, Zhigang ZHAO1, Ling JIANG1, Jianmin WAN1,*( )
)
Received:2018-03-13
Revised:2018-05-29
Online:2019-01-10
Published:2019-01-10
Contact:
Jianmin WAN
摘要:
【目的】叶色突变相关基因的鉴定与克隆为研究叶绿体发育、叶绿素合成和光合作用等分子机制提供理论基础。【方法】从常规粳稻镇糯19杂交后代中分离出一个低温移栽后叶色转成白条纹的自然变异突变体, 命名为wltt (white stripe leaf after transplanting at low temperature)。成熟期测定野生型和wltt的主要农艺性状,分别在苗期、移栽后15 d和同时期直播条件下测定新生叶片的色素含量并观察叶绿体的超微结构;将wltt和野生型正反交进行遗传分析;用wltt与籼稻9311杂交产生的F2作为定位群体进行基因定位;采用RT-qPCR 分析叶绿体发育、叶绿素合成和光合作用相关基因在野生型和wltt中的表达水平。【结果】wltt突变体在苗期表现正常绿色,移栽15 d后心叶出现白条纹叶表型,至分蘖末期心叶叶色恢复;而不经移栽,突变体不会出现白条纹叶。人工模拟实验表明该表型是由低温条件下根损伤引起的。与野生型相比,wltt突变体移栽后的新生叶色素含量显著降低,光合速率下降;同时株高变矮,穗长、剑叶长和每穗粒数均显著降低。叶绿体的超微结构显示,突变体的叶肉细胞中,仅少数细胞含有正常的叶绿体,其余大部分叶肉细胞不含叶绿体。进一步研究发现,突变体中部分光合系统相关基因和叶绿体发育相关基因表达下调,叶绿素生物合成相关的14个基因表达也下调。遗传分析表明,该突变性状受一对隐性核基因控制。利用wltt突变体/9311的F2群体,将该基因定位于水稻第2染色体着丝粒附近853 kb区间内。目前,该区间内没有叶色相关基因的报道。【结论】WLTT是低温条件下移栽调控叶片转色的关键基因,在叶绿体发育过程中发挥重要作用。
中图分类号:
林添资, 孙立亭, 龚红兵, 王益华, 刘玲珑, 赵志刚, 江玲, 万建民. 一个水稻低温移栽白条纹突变体wltt的鉴定和基因定位[J]. 中国水稻科学, 2019, 33(1): 1-11.
Tianzi LIN, Liting SUN, Hongbin GONG, Yihua WANG, Linglong LIU, Zhigang ZHAO, Ling JIANG, Jianmin WAN. Identification and Gene Mapping of a white-stripe leaf after transplanting at low temperature Mutant in Rice[J]. Chinese Journal OF Rice Science, 2019, 33(1): 1-11.
| 引物名称 Primer name | 前引物 Forward sequence (5′-3′) | 后引物 Reverse sequence (5′-3′) | BAC克隆 BAC clone |
|---|---|---|---|
| L1 | ATTCAGTAAGACTACACGCAT | AATGACAGATTACTTGTTCCA | OJ1756_H07 |
| L6 | CTAACATAATGGGTAAAGAGG | TTAGTTGGTTGCCGTGT | OJ1124_E11 |
| L8 | ATAGTTTAGGGAGTTATGTGCT | CGTGTGCCTATTGACTTCTC | OSJNBa0030M21 |
| L11 | ACAGAACGGAACGGGATA | CTCACAATCTTTTATCACCCA | OSJNBa0078K05 |
| L14 | AACCAAGAATCGGAAAGAA | ATCCCATTTCCATTTCTCT | OSJNBa0008C07 |
| L16 | TTTCCTGAGCGAATCCA | AAAAGGCACTTATGAGACACT | OSJNBb0080M22 |
| L18 | TAGGTGGTTGAATGGTGC | TATGCTTCTTTTGGGTTG | P0543C11 |
| L20 | TGAGATACGCAGAATGGG | GAGGAGGATGCAGGGAC | P0705A04 |
| L22 | GTTCTTTTGTCTTCCCTCA | ATTATCCTTGGTCTTGGTAT | OJ1134_B09 |
| L26 | TTGGAGAATGAAGTTGCTAA | TTACCAAGCAGGACTAAAGAT | OSJNBb0037J12 |
| I2-7 | GAACCAGTCCGCTCTCTGAC | TACGCGTCGTGTATCGTAGC | OSJNBa0035A24 |
表1 基因定位引物
Table 1 Primers used for gene mapping.
| 引物名称 Primer name | 前引物 Forward sequence (5′-3′) | 后引物 Reverse sequence (5′-3′) | BAC克隆 BAC clone |
|---|---|---|---|
| L1 | ATTCAGTAAGACTACACGCAT | AATGACAGATTACTTGTTCCA | OJ1756_H07 |
| L6 | CTAACATAATGGGTAAAGAGG | TTAGTTGGTTGCCGTGT | OJ1124_E11 |
| L8 | ATAGTTTAGGGAGTTATGTGCT | CGTGTGCCTATTGACTTCTC | OSJNBa0030M21 |
| L11 | ACAGAACGGAACGGGATA | CTCACAATCTTTTATCACCCA | OSJNBa0078K05 |
| L14 | AACCAAGAATCGGAAAGAA | ATCCCATTTCCATTTCTCT | OSJNBa0008C07 |
| L16 | TTTCCTGAGCGAATCCA | AAAAGGCACTTATGAGACACT | OSJNBb0080M22 |
| L18 | TAGGTGGTTGAATGGTGC | TATGCTTCTTTTGGGTTG | P0543C11 |
| L20 | TGAGATACGCAGAATGGG | GAGGAGGATGCAGGGAC | P0705A04 |
| L22 | GTTCTTTTGTCTTCCCTCA | ATTATCCTTGGTCTTGGTAT | OJ1134_B09 |
| L26 | TTGGAGAATGAAGTTGCTAA | TTACCAAGCAGGACTAAAGAT | OSJNBb0037J12 |
| I2-7 | GAACCAGTCCGCTCTCTGAC | TACGCGTCGTGTATCGTAGC | OSJNBa0035A24 |
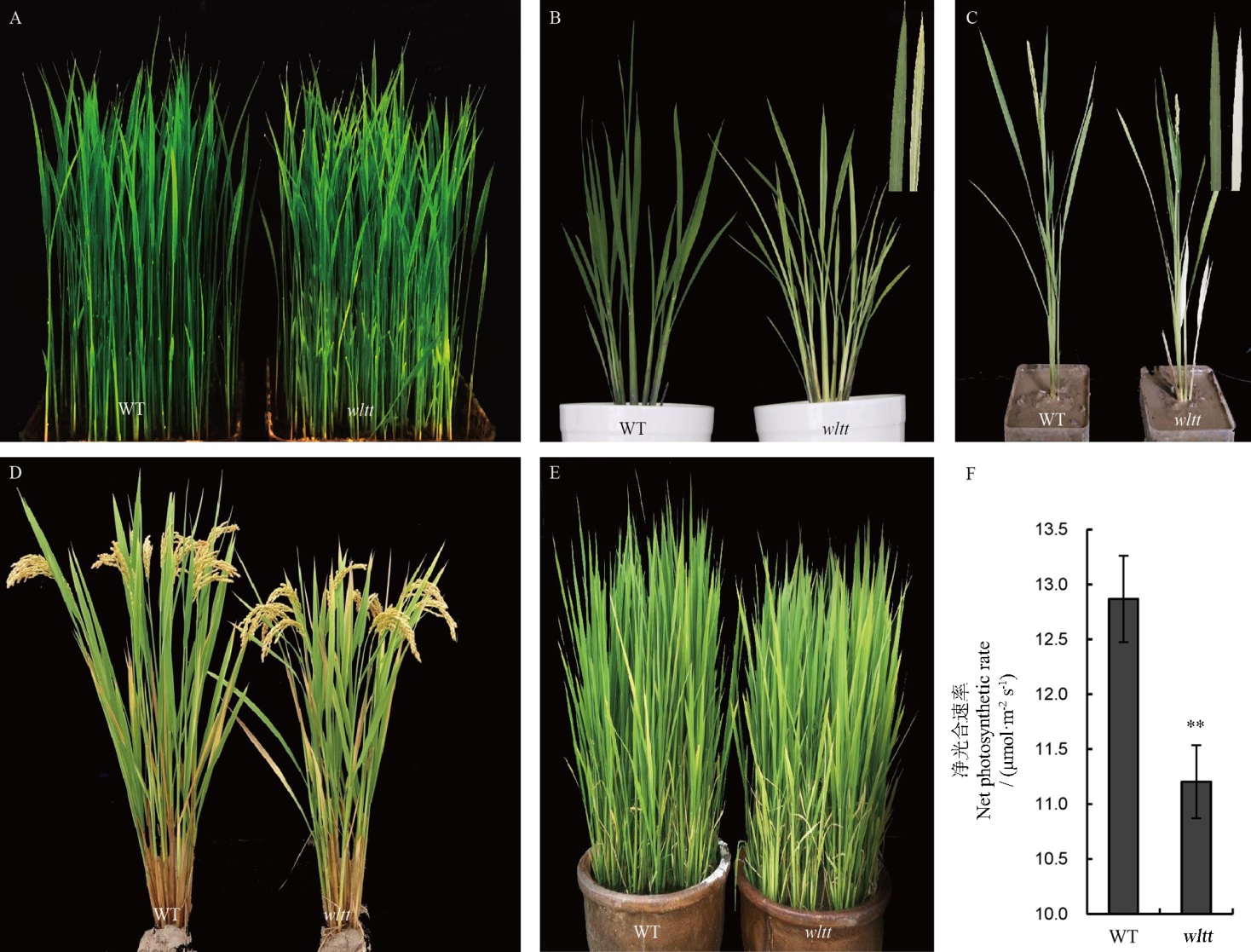
图1 野生型与wltt突变体不同时期的表型A-苗期;B-分蘖期,移栽后15 d,右上角为野生型(左)和突变体(右)的新完全展开叶;C-抽穗期移栽后15 d,右上角为野生型(左)和突变体(右)的再生分蘖的叶片;D-6月10日正常移栽的成熟期植株;E-直播条件下分蘖期的植株;F-正常移栽分蘖期植株新生叶片的净光合速率。WT-野生型。**表示经t测验后,P<0.01下差异显著。
Fig. 1. Phenotypic characterization of the wltt mutant and its wild type(WT). A, Seedling stage; B, Fifteen days after transplanting, at the tillering stage; The insert represents the new fully-expanded leaf blades of the wild type (left) and the mutant (right); C, Fifteen days after transplanting at the heading stage. The inserts represents the leaves of regenerated tillers of the wild type (left) and the mutant (right); D, Plants at the mature stage (transplanting at the tillering stage). E, Wild-type and mutant plants at the tillering stage under direct seeding. F, Net photosynthetic rate of new fully expanded leaf blades of the wild type and wltt mutant at the tillering stage. WT, Wild type. ** P<0.01(Student’s t test).
| 材料 Material | 株高 Plant height /cm | 有效穗数 No. of effective panicles | 剑叶长 Flag leaf length /cm | 穗长 Panicle length /cm | 每穗总粒数 No. of spikelets per panicle | 结实率 Seed-setting rate /% | 千粒重 1000-grain weight /g | |
|---|---|---|---|---|---|---|---|---|
| WT | 86.7±0.2 | 9.2±0.4 | 22.28±0.53 | 20.44±0.46 | 173.6±5.7 | 95.8±0.4 | 29.18±0.26 | |
| wltt | 75.8±0.5** | 8.2±0.4 | 18.10±0.37** | 18.40±0.41** | 136.2±6.9** | 95.6±1.7 | 28.46±0.32 | |
表2 野生型与突变体wltt的主要农艺性状比较
Table 2 Comparison of major agronomic traits between the wltt mutant and its wild type(WT).
| 材料 Material | 株高 Plant height /cm | 有效穗数 No. of effective panicles | 剑叶长 Flag leaf length /cm | 穗长 Panicle length /cm | 每穗总粒数 No. of spikelets per panicle | 结实率 Seed-setting rate /% | 千粒重 1000-grain weight /g | |
|---|---|---|---|---|---|---|---|---|
| WT | 86.7±0.2 | 9.2±0.4 | 22.28±0.53 | 20.44±0.46 | 173.6±5.7 | 95.8±0.4 | 29.18±0.26 | |
| wltt | 75.8±0.5** | 8.2±0.4 | 18.10±0.37** | 18.40±0.41** | 136.2±6.9** | 95.6±1.7 | 28.46±0.32 | |
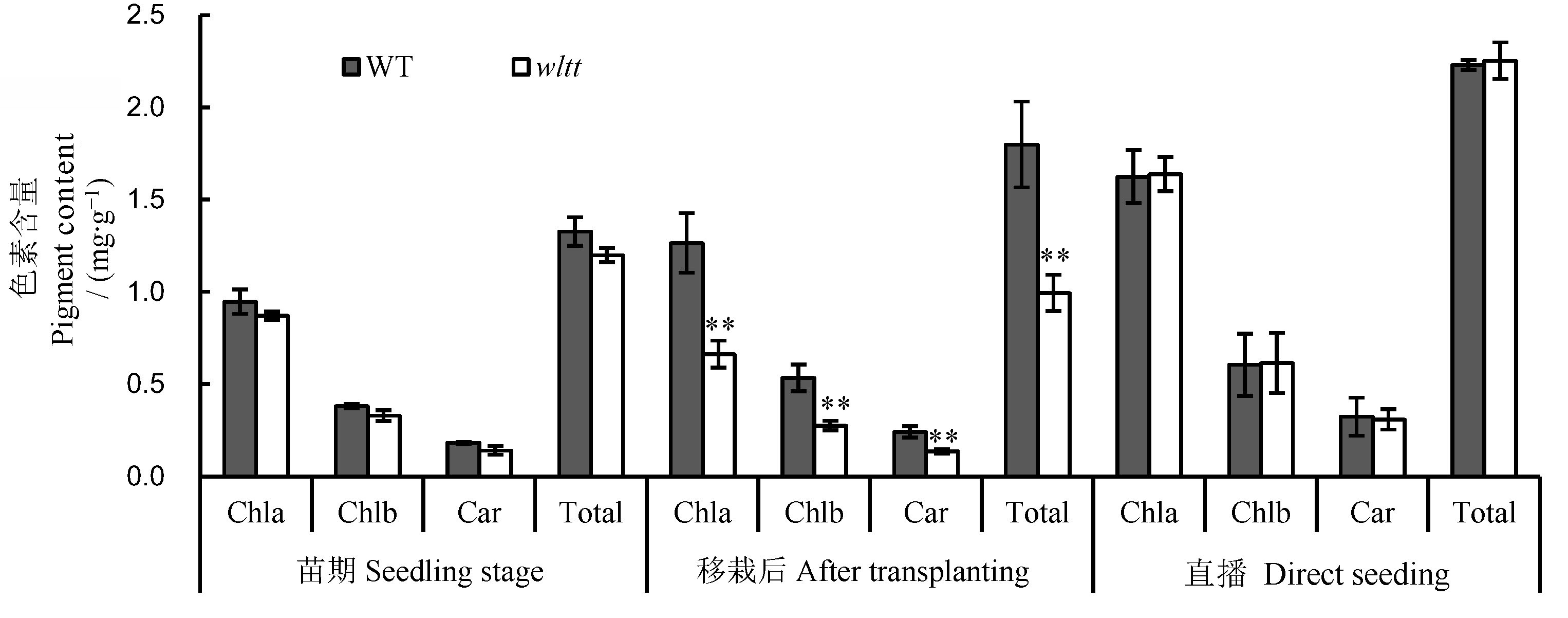
图2 wltt突变体和野生型叶片色素含量分析 Chla-叶绿素a;Chlb-叶绿素b;Car-类胡萝卜素;Total-总色素含量。误差线表示3次独立实验的标准差。**表示野生型与突变体间差异达0.01显著水平(t测验)。
Fig. 2. Pigment contents in leaves of wltt mutant and its wild type(WT). Chla, Chlorophyll a; Chlb, Chlorophyll b; Car, Carotenoids; Total, Total pigment contents. Values are presented as mean±SD. **The difference between the wild type and wltt is significant at 0.01 level according to Student’s t test.
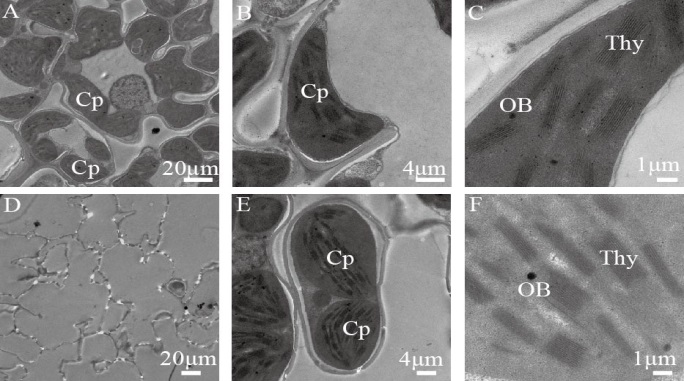
图3 野生型和突变体wltt的叶绿体超微结构 A~C为野生型的叶绿体超微结构;D~F为突变体wltt中同时含有异常结构的叶绿体(D)和正常结构的叶绿体(E~F)。Cp-叶绿体; Thy-类囊体; OB-嗜锇体。
Fig. 3. Transmission electron microscopic (TEM) images of chloroplast ultrastructure in the wild type and the wltt mutant. A-C, Transmission electron microscopic (TEM) images of chloroplast ultrastructure in wild type; D-F, Two types of chloroplasts with (E, F) or without (D) a normal ultrastructure in the wltt mutant; Cp, Chloroplast; Thy, Thylakoid lamellae; OB, Osmiophilic body.

图4 叶绿体发育及光合系统相关基因在野生型和wltt突变体中的表达分析误差线表示3次独立实验的标准差。**表示野生型与突变体间差异达0.05和0.01显著水平(t测验)。
Fig. 4. Expression analysis of genes associated with chloroplast development and photosynthetic system in the wild type(WT) and wltt mutant. Mean±SD (n=3). **The difference between the wild type and wltt is significant at 0.01 level according to Student’s t test.

图5 叶绿素合成相关基因在野生型和突变体wltt中的表达分析误差线表示3次独立实验的标准差。*和**分别表示野生型与突变体间差异达0.05和0.01显著水平(t测验)。
Fig. 5. Expression analysis of genes associated with chlorophyll biogenesis in the wild type(WT) and wltt mutant. Means ± SD (n=3). * and **The difference between the wild type and wltt is significant at 0.05 and 0.01 level, respectively according to Student’s t test.
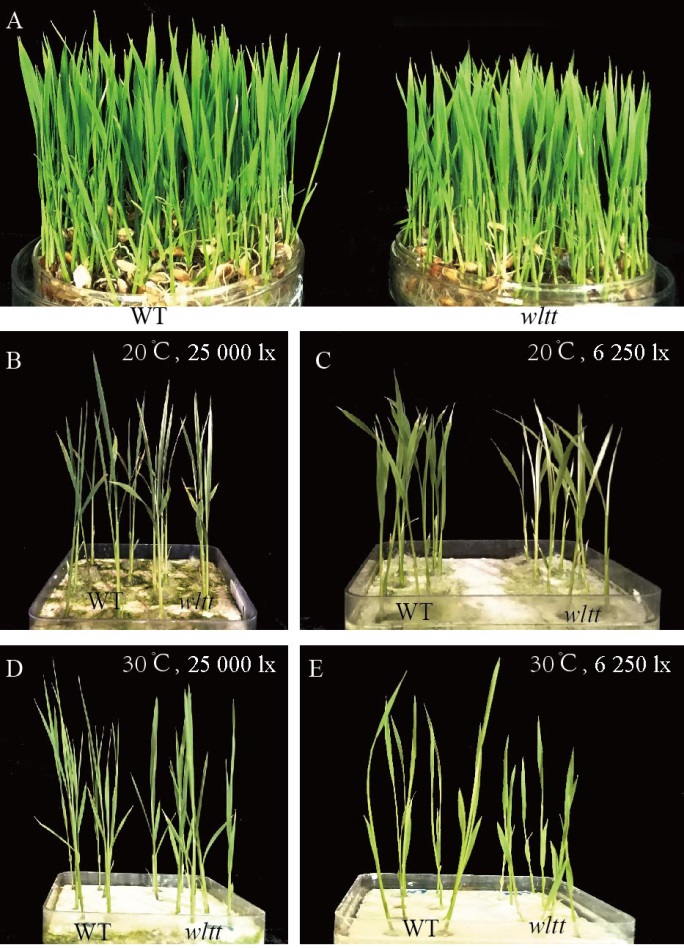
图6 环境条件对叶色变异的影响 A–不进行剪根处理;B, C-剪根处理10 d后,20℃条件下,光照强度为25 000 lx(B)和6250 lx(C);D, E-剪根处理10 d后,30℃条件下,光照强度为25 000 lx(D)和6250 lx(E)。
Fig. 6. Effects of environmental conditions on leaf color variation. A, The seedlings of the wild type and wltt with non-injured roots; B, C, The seedlings of wild type and wltt 10 days after root cutting treatment at 20℃ at the light intensity of 25 000 lx(B) and 6250 lx(C), respectively. D, E, The seedlings of wild type and wltt 10 days after root cutting treatment at 30℃ at the light intensity of 25 000 lx(D) and 6250 lx(E), respectively.
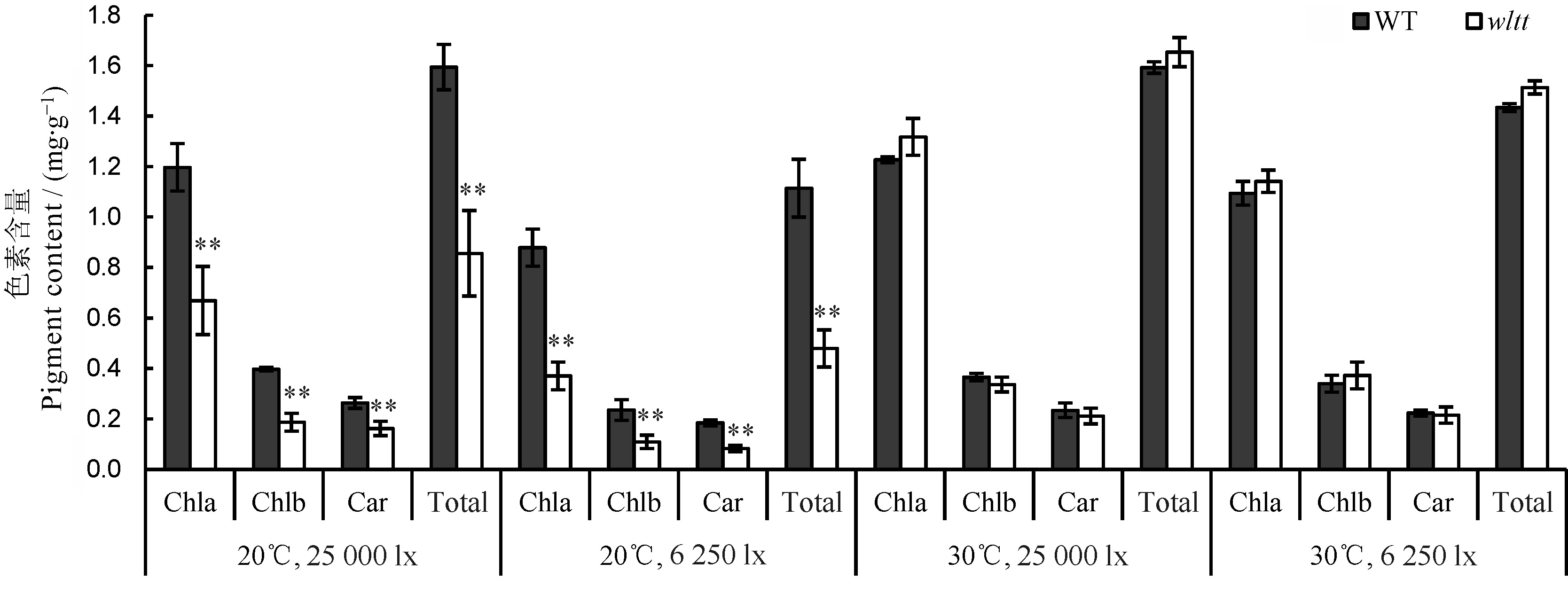
图7 环境条件对剪根处理后色素含量的影响误差线表示3次独立实验的标准差。**表示野生型与突变体间差异达0.01显著水平(t测验)。
Fig. 7. Effects of environmental conditions on pigment contents after root cutting treatment. Means±SD (n=3). **The difference between the wild type and wltt is significant at 0.01 level according to Student’s t test.
| 杂交组合 Cross | 正常株数 No. of normal plants | 白条纹株数 No. of white stripe plants | 实际分离比 Segregation ratio | χ2(3:1) |
|---|---|---|---|---|
| WT×wltt | 375 | 127 | 2.95:1 | 0.02 |
| wltt×WT | 368 | 130 | 2.83:1 | 0.06 |
表3 野生型与wltt突变体的F2代分离比
Table 3 Segregation of F2 population from wltt mutant and its wild type(WT).
| 杂交组合 Cross | 正常株数 No. of normal plants | 白条纹株数 No. of white stripe plants | 实际分离比 Segregation ratio | χ2(3:1) |
|---|---|---|---|---|
| WT×wltt | 375 | 127 | 2.95:1 | 0.02 |
| wltt×WT | 368 | 130 | 2.83:1 | 0.06 |

图8 WLTT在第2染色体上的定位 A–基因位点初定位在第2染色体着丝粒附近InDel标记I2-5和I2-8之间;B-基因位点精细定位到标记L22和L26之间,物理距离853 kb。n为定位所用的白条纹叶植株数。
Fig. 8. Location of WLTT on rice chromosome 2. A, The gene is mapped to the centromeric region of chromosome 2 between the InDel markers I2-5 and I2-8; B, Mapping of the gene locus between markers L22 and L26 within an 853 kb region. CEN, Centromere; n, Number of individuals with white stripe leaf after transplanting.
| [1] | Leister D.Chloroplast research in the genomic age.Trends Genet, 2003, 19(1): 47-56. |
| [2] | Kong W, Yu X, Chen H, Liu L, Xiao Y, Wang Y, Wang C, Lin Y, Yu Y, Wang C, Jiang L, Zhai H, Zhao Z, Wan J.The catalytic subunit of magnesium-protoporphyrin IX monomethyl ester cyclase forms a chloroplast complex to regulate chlorophyll biosynthesis in rice.Plant Mol Biol, 2016, 92(1-2): 177-191. |
| [3] | Wang L, Wang C, Wang Y, Niu M, Ren Y, Zhou K, Zhang H, Lin Q, Wu F, Cheng Z, Wang J, Zhang X, Guo X, Jiang L, Lei C, Wang J, Zhu S, Zhao Z, Wan J.WSL3, a component of the plastid-encoded plastid RNA polymerase, is essential for early chloroplast development in rice.Plant Mol Biol, 2016, 92(4/5): 581-595. |
| [4] | Zhang Z, Tan J, Shi Z, Xie Q, Xing Y, Liu C, Chen Q, Zhu H, Wang J, Zhang J, Zhang G. Albino Leaf1 that encodes the sole octotricopeptide repeat protein is responsible for chloroplast development. Plant Physiol, 2016, 171(2): 1182-1191. |
| [5] | Fambrini M, Castagna A, Vecchia F D, Degl Innocenti E, Ranieri A, Vernieri P, Pardossi A, Guidi L, Rascio N, Pugliesi C.Characterization of a pigment-deficient mutant of sunflower (Helianthus annuus L.) with abnormal chloroplast biogenesis, reduced PSII activity and low endogenous level of abscisic acid. Plant Breeding, 2004, 6: 645-650. |
| [6] | Agrawal G K, Yamazaki M, Kobayashi M, Hirochika R, Miyao A, Hirochika H.Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel ostatc gene. Plant Physiol, 2001, 125(3): 1248-1257. |
| [7] | Parks B M, Quail P H.Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell, 1991, 3(11): 1177-1186. |
| [8] | Su N, Hu M L, Wu D X, Wu F Q, Fei G L, Lan Y, Chen X L, Shu X L, Zhang X, Guo X P, Cheng Z J, Lei C L, Qi C K, Jiang L, Wang H, Wan J M.Disruption of a rice pentatricopeptide repeat protein causes a seedling- specific albino phenotype and its utilization to enhance seed purity in hybrid rice production.Plant Physiol, 2012, 159(1): 227-238. |
| [9] | 谭炎宁, 孙学武, 袁定阳, 孙志忠, 余东, 何强, 段美娟, 邓华凤, 袁隆平. 水稻单叶独立转绿型黄化突变体grc2 的鉴定与基因精细定位. 作物学报, 2015, 41(6): 831-837. |
| Tan Y N, Sun X W, Yuan D Y, Sun Z Z, Yu D, He Q, Duan M J, Deng H F, Yuan L P.Identification and fine mapping of green-revertible chlorina gene grc2 in rice(Oryza sativa L.). Acta Agron Sin, 2015, 41(6): 831-837. (in Chinese with English abstract) | |
| [10] | 钱前, 朱旭东, 曾大力, 张小惠, 严学强, 熊振民. 细胞质基因控制的新特异材料白绿苗的研究. 作物品种资源, 1996(4): 11-12. |
| Qian Q, Zhu X D, Zeng D L, Zhang X H, Yan X Q, Xiong Z M.The study on a new special material, white-green rice which controlled by plasma gene.J Crop Resour, 1996(4): 11-12. (in Chinese). | |
| [11] | 李贤勇, 王楚桃, 李顺武, 何永歆, 陈世全. 一个水稻高叶绿素含量基因的发现. 西南农业学报, 2002, 15(4): 122-123. |
| Li X Y, Wang C T, Li S W, He Y X, Chen S Q.The discovery of a high chlorophyll content gene in rice.Southwest China J Agric Sci, 2002, 15(4): 122-123. (in Chinese with English abstract) | |
| [12] | Lee S, Kim J H, Yoo E S, Lee C H, Hirochika H, An G.Differential regulation of chlorophyll a oxygenase genes in rice.Plant Mol Biol, 2005, 57(6): 805-818. |
| [13] | Yang Y L, Xu J, Huang L C, Leng Y J, Dai L P, Rao Y C, Chen L, Wang Y Q, Tu Z J, Hu J, Ren D Y, Zhang G H, Zhu L, Guo L B, Qian Q, Zeng D L. PGL , encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J Exp Bot, 2016, 67(5): 1297-1310. |
| [14] | Kusumi K, Yara A, Mitsui N, Tozawa Y, Iba K.Characterization of a rice nuclear-encoded plastid RNA polymerase gene OsRpoTp. Plant Cell Physiol, 2004, 45(9): 1194-1201. |
| [15] | Sugimoto H, Kusumi K, Tozawa Y, Yazaki J, Kishimoto N, Kikuchi S, Iba K.The virescent-2 mutation inhibits translation of plastid transcripts for the plastid genetic system at an early stage of chloroplast differentiation. Plant Cell Physiol, 2004, 45(8): 985-996. |
| [16] | Beale S I.Green genes gleaned.Trends Plant Sci, 2005, 10(7): 309-312. |
| [17] | Nagata N, Tanaka R, Satoh S, Tanaka A.Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell, 2005, 17(1): 233-240. |
| [18] | Goh C H, Satoh K, Kikuchi S, Kim S C, Ko S M, Kang H G, Jeon J S, Kim C S, Park Y.Mitochondrial activity in illuminated leaves of chlorophyll-deficient mutant rice OsCHLH seedlings. Plant Biotechnol Rep, 2010, 4(4): 281-291. |
| [19] | Zhang H T, Li J J, Yoo J H, Yoo S C, Cho S H, Koh H J, Seo H S, Paek N C.Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol Biol, 2006, 62(3): 325-337. |
| [20] | Wang P R, Gao J X, Wan C M, Zhang F T, Xu Z J, Huang X Q, Sun X Q, Deng X J.Divinyl chlorophyll(ide) a can be converted to monovinyl chlorophyll(ide) a by a |
| divinyl reductase in rice.Plant Physiol, 2010, 153(3): 994-1003. | |
| [21] | Sakuraba Y, Rahman M L, Cho S H, Kim Y S, Koh H J, Yoo S C, Paek N C.The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions.Plant J, 2013, 74(1): 122-133. |
| [22] | Yang Q S, He H, Li H Y, Tian H, Zhang J J, Zhai L G, Chen J D, Wu H, Yi G J, He Z H, Peng X X.NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and rubisco formation in rice.PLoS ONE, 2011, 6(5): e20015. |
| [23] | Wu Z M, Zhang X, He B, Diao L P, Sheng S L, Wang J L, Guo X P, Su N, Wang L F, Jiang L, Wang C M, Zhai H Q, Wan J M.A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis.Plant Physiol, 2007, 145(1): 29-40. |
| [24] | Yagi Y, Ishizaki Y, Nakahira Y, Tozawa Y, Shiina T.Eukaryotic-type plastid nucleoid protein pTAC3 is essential for transcription by the bacterial-type plastid RNA polymerase.Proc Natl Acad Sci USA, 2012, 109(19): 7541-7546. |
| [25] | Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustun S, Melzer M, Petersen K, Lein W, Bornke F Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: Evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell, 2010, 22: 1498-1515. |
| [26] | Wang Y, Wang C, Zheng M, Lyu J, Xu Y, Li X, Niu M, Long W, Wang D, Wang H Y, William T, Wang Y, Wan J.WHITE PANICLE1, a Val-tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development.Plant Physiol, 2016, 170(4): 2110-2123. |
| [27] | Wu L, Wu J, Liu Y, Gong X, Xu J, Lin D, Dong Y.The rice pentatricopeptide repeat gene TCD10 is needed for chloroplast development under cold stress. Rice, 2016, 9: 67. |
| [28] | Tang J, Zhang W, Wen K, Chen G, Sun J, Tian Y, Tang W, Yu J, An H, Wu T, Kong F, Terzaghi W, Wang C, Wan J.OsPPR6, a pentatricopeptide repeat protein involved in editing and splicing chloroplast RNA, is required for chloroplast biogenesis in rice.Plant Mol Biol, 2017, 95(4/5): 345-357. |
| [29] | Yue R, Wang X, Chen J, Ma X, Zhang H, Mao C, Wu P.A rice stromal processing peptidase regulates chloroplast and root development.Plant Cell Physiol, 2010, 51(3): 475-485. |
| [30] | Dong H, Fei G L, Wu C Y, Wu F Q, Sun Y Y, Chen M J, Ren Y L, Zhou K N, Cheng Z J, Wang J L, Jiang L, Zhang X, Guo X P, Lei C L, Su N, Wang H, Wan J M.A rice Virescent-Yellow Leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiol, 2013, 162(4): 1867-1880. |
| [31] | Zhou S, Sawicki A, Willows R D, Luo M.C-terminal residues of Oryza sativa GUN4 are required for the activation of the ChlH subunit of magnesium chelatase in chlorophyll synthesis. FEBS Lett, 2012,586(3): 205-210. |
| [32] | Yoshida S, Forno D A, Cock J A H, Gomez K A. Laboratory Manual for Physiological Studies of Rice. Los Banos,Philippines: The International Rice Research Institute, 1976: 61. |
| [33] | McCouch S R, Kochert G, Yu Z H, Wang Z Y, Khush G S, Coffman W R, Tanksley S D. Molecular mapping of rice chromosome.Theor Appl Genet, 1998, 76: 815-829. |
| [34] | Livak K J, Schmittgen T D.Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) method.Methods, 2001, 25(4): 402-408. |
| [35] | Shi X, Chen S, Peng Y, Wang Y, Chen J, Hu Z, Wang B, Li A, Chao D, Li Y, Teng S.TSC1 enables plastid development under dark conditions, contributing to rice adaptation to transplantation shock.J Integr Plant Biol, 2018, 60(2): 112-129. |
| [36] | Kensuke K, Shoko H, Hiroshi S, Yoko C, Osanu M, Koh I.Contribution of chloroplast biogenesis to carbon- nitrogen balance during early leaf development in rice.J Plant Res, 2010, 123(4): 617-622. |
| [37] | Hiroki S, Kensuke K, Ko N, Masahiro Y, Atsushi Y, Koh I.The rice nuclear gene, VIRESCENT 2 , is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J, 2007, 52(3): 512-527. |
| [38] | Gong X D, Su Q Q, Lin D Z, Jiang Q, Xu J L, Zhang J H, Teng S, Dong Y J.The rice OsV4 encoding a novel pentatricopeptide repeat protein is required for chloroplast development during the early leaf stage under cold stress. J Integr Plant Biol, 2014, 56(4): 400-410 |
| [39] | Jiang Q, Mei J, Gong X D, Xu J L, Zhang J H, Teng S, Lin D Z, Dong Y J. Importance of the rice TCD9 encoding subunit of chaperonin protein 60(Cpn60)for the chloroplast development during the early leaf stage. Plant Sci, 2014, 215/216: 172-179. |
| [40] | Song J, Wei X J, Shao G N, Sheng Z H, Chen D B, Liu C L, Jiao G A, Xie L L, Tang S Q, Hu P S.The rice nuclear gene WLP1 encoding a chloroplast ribosome L13 protein is needed for chloroplast development in rice grown under low temperature conditions. Plant Mol Biol, 2014, 84(3): 301-314 |
| [41] | Pakrasi H B.Genetic analysis of the form and function of photosystem Ⅰ and photosystem Ⅱ.Annu Rev Genet, 1995, 29: 755-776. |
| [42] | Boudreau E, Takahashi Y, Lemieux C, Turmel M, Rochaix J D.The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J, 1997, 16(20): 6095-6104. |
| [43] | Santis-Maciossek G D, Kofer W, Bock A, Schoch S, Maier R M, Wanner G, Rüdiger W, Hans-Ulrich K, Herrmann R G. Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: Molecular biology biochemistry and ultrastructure. Plant J, 1999, 18: 477-489. |
| [44] | Rogalski M, Ruf S, Bock R.Tobacco plastid ribosomal protein S18 is essential for cell survival.Nucl Acids Res, 2006, 34: 4537-4545. |
| [45] | Fleischmann T T, Scharff L B, Alkatib S, Hasdorf S, Schottler M A, Bock R.Nonessential plastid-encoded ribosomal proteins in tobacco: A developmental role for plastid translation and implications for reductive genome evolution.Plant Cell, 2011, 23(9): 3137-3155. |
| [46] | 夏家平, 郭会君, 谢永盾, 赵林姝, 古佳玉, 赵世荣, 李军辉, 刘录祥. 小麦叶绿素缺失突变体Mt135的叶绿体基因差异表达分析. 中国水稻科学, 2012, 38(11):2122-2130. |
| Xia J P, Guo H J, Xie Y D, Zhao L S, Gu J Y, Zhao S R, Li J H, Liu L X.Differential expression of chloroplast genes in chlorophyll-deficient wheat mutantMt135 derived from space mutagenesis. Chin J Rice Sci, 2012(11): 2122-2130. | |
| [47] | Chen T, Zhang Y, Zhao L, Zhu Z, Lin J, Zhang S, Wang C.Fine mapping and candidate gene analysis of a green-revertible albino gene gra(t) in rice. J Genet Genomics, 2009, 36(2): 117-123. |
| [48] | 李燕群, 钟萍, 高志艳, 朱柏羊, 陈丹, 孙昌辉, 王平荣, 邓晓建. 水稻斑马叶突变体zebra524的表型鉴定及候选基因分析. 中国农业科学, 2014, 47(15): 2907-2915. |
| Li Y Q, Zhong P, Gao Z Y, Zhu B Y, Chen D, Sun C H, Wang P R, Deng X J.Morphological characterization and candidate gene analysis of zebra leaf mutant zebra524 in rice. Sci Agric Sin, 2014, 47(15): 2907-2915. | |
| [49] | Lin D, Jiang Q, Zheng K, Chen S, Zhou H, Gong X, Xu J, Teng S, Dong Y.Mutation of the rice ASL2 gene encoding plastid ribosomal protein L21 causes chloroplast developmental defects and seedling death. Plant Biol (Stuttg), 2015, 17(3): 599-607. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||