
中国水稻科学 ›› 2018, Vol. 32 ›› Issue (5): 427-436.DOI: 10.16819/j.1001-7216.2018.7037
张占田, 孙雅菲, 艾昊, 罗闻真, 冯冰, 孙文献, 徐国华, 孙淑斌*( )
)
收稿日期:2017-03-30
修回日期:2017-08-18
出版日期:2018-09-10
发布日期:2018-09-10
通讯作者:
孙淑斌
基金资助:
Zhantian ZHANG, Yafei SUN, Hao AI, Wenzhen LUO, Bing FENG, Wenxian SUN, Guohua XU, Shubin SUN*( )
)
Received:2017-03-30
Revised:2017-08-18
Online:2018-09-10
Published:2018-09-10
Contact:
Shubin SUN
摘要:
【目的】水稻OsSHR2(LOC_Os03g31880)基因为拟南芥AtSHR的同源基因,与OsSHR1、OsSCR1和OsSCR2 同属于水稻GRAS转录因子家族。已有研究报道,转录因子基因SHR和SCR共同调控植物根系、叶片的发育,并参与各项生命活动。本研究旨在阐明OsSHR2在水稻中的时空表达特征及其在营养生长中的调控作用。【方法】通过生物信息学分析、表达模式分析、萌发动力学分析和水培实验验证该基因的功能。【结果】生物信息学分析发现OsSHR2、OsSHR1、OsSCR1和OsSCR2与拟南芥和其他物种的SHR亚家族和SCR亚家族成员具有很高的序列一致性;表达模式和pOsSHR2::GUS材料染色分析发现,OsSHR2在整个生长发育过程中的根系、叶片、维管组织和生殖器官中表达强烈,并集中在根尖的中柱、侧根原基和叶片及茎维管组织的中心表达,在野生型的地上部和根系中,OsSHR2受缺磷影响下调表达;对获得的OsSHR2的CRISPR-Cas9突变体osshr2进行种子萌发实验和水培实验,发现与野生型相比,osshr2的萌发时间延后,萌发率降低,在正常供磷和缺磷处理下,osshr2的地上部和根系长度显著小于野生型。【结论】OsSHR2在地上部和根系的发育、维管组织形成以及营养与生殖生长中具有重要作用,这为今后OsSHR2在分子育种等领域的应用奠定理论基础。
中图分类号:
张占田, 孙雅菲, 艾昊, 罗闻真, 冯冰, 孙文献, 徐国华, 孙淑斌. 水稻转录因子基因OsSHR2的表达特征及其在营养生长中的调控作用[J]. 中国水稻科学, 2018, 32(5): 427-436.
Zhantian ZHANG, Yafei SUN, Hao AI, Wenzhen LUO, Bing FENG, Wenxian SUN, Guohua XU, Shubin SUN. Expression Patterns and Regulation of Transcription Factor Gene OsSHR2 in Vegetative Growth in Rice[J]. Chinese Journal OF Rice Science, 2018, 32(5): 427-436.
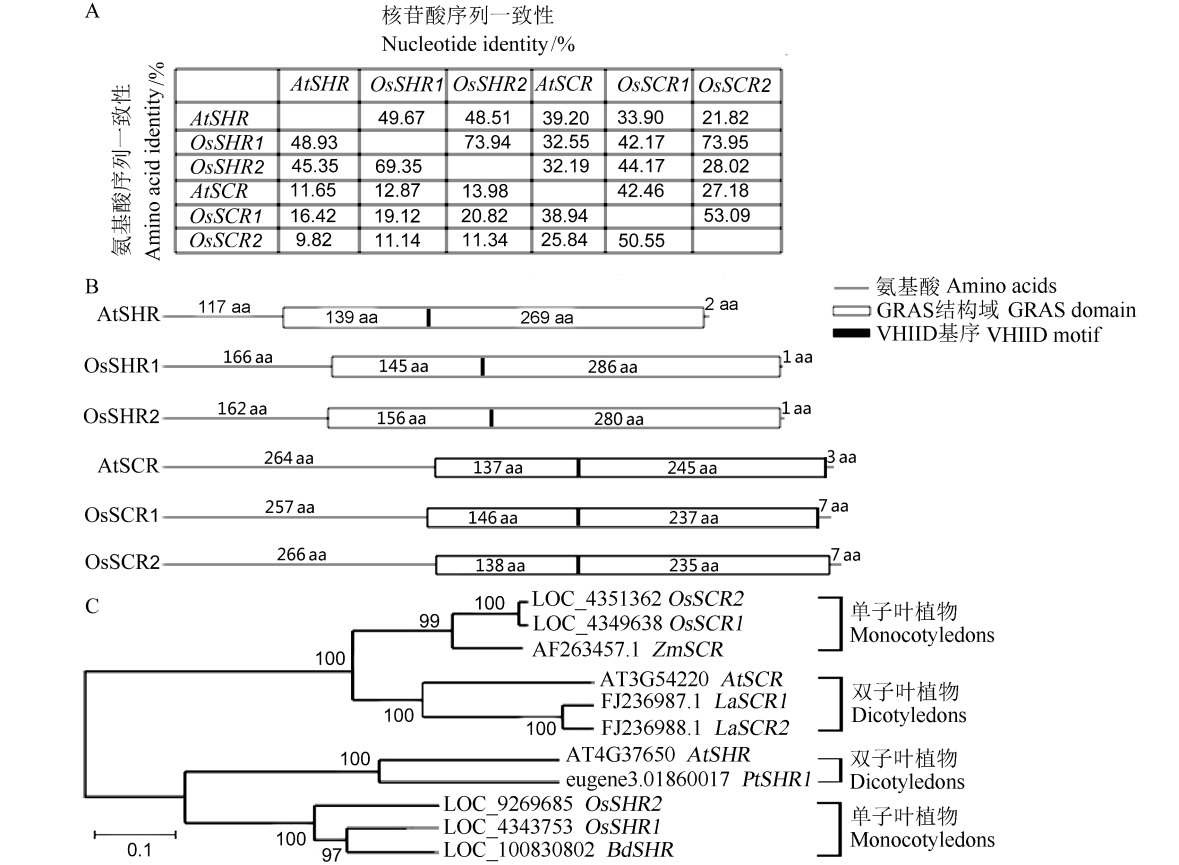
图1 不同物种SHR和SCR同源基因的生物信息学分析 A–拟南芥和水稻的SHR和SCR同源基因氨基酸和核苷酸的序列一致性分析;B–通过基因家族分析网站InterPro(http://www.ebi.ac.uk/ interpro/)进行拟南芥与水稻的SHR和SCR同源基因GRAS保守结构域和保守基序VHIID的位置预测;C–不同物种SHR和SCR同源基因的进化树分析。
Fig. 1. Bioinformatics analysis of SHR and SCR homologous genes of different species. A, Sequence homology analysis of amino acids and nucleic acid of SHR and SCR homologous genes in Arabidopsis thaliana and rice; B, Prediction of the positions of SHR and SCR homologous GRAS conserved domains and conserved motifs in Arabidopsis thaliana and rice by InterPro (http://www.ebi.ac.uk/interpro/); C, Phylogenetic tree analysis of SHR and SCR homologous genes of different species.
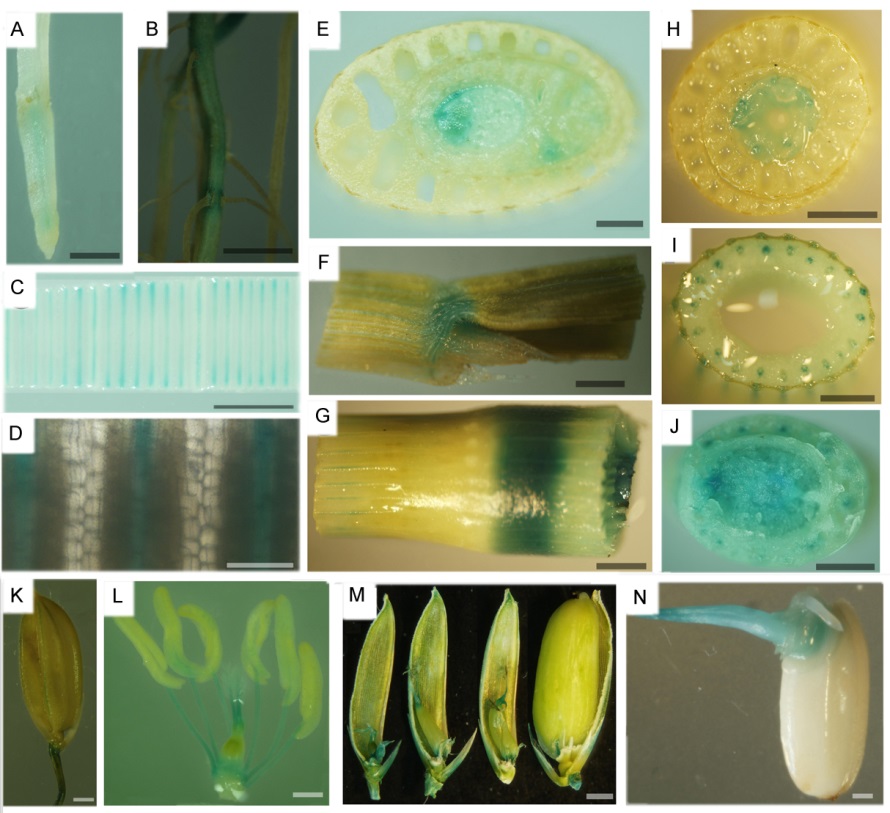
图3 pOsSHR2::GUS材料的GUS染色鉴定 A–种子根;B–种子根及侧根;C–新叶;D–新叶的放大图;E–叶原基;F–叶舌;G–茎节;H–茎及叶鞘;I–茎;J–茎基部;K–颖壳及小穗轴;L–颖花;M–子房和柱头;N–胚(萌发后3 d)。A~E,J:水稻苗期;F,G,I,K~N:水稻灌浆期;H:分蘖期。A~J中标尺为2 mm;K~N中标尺为0.5 mm。
Fig. 3. Identification of OsSHR2 promoter-driven tissue-specific GUS staining. A, Seed root; B, Seed root and lateral root; C, Young leaf; D, The enlarged view of the young leaf; E, Leaf primordium; F, Ligule; G, Node; H, Stem and leaf sheath; I, Stem; J, Basal stem; K, Husk and rachilla; L, Spikelet; M, Ovary and stigma; N, Embryo(three days after germination). A-E, J: Seedling stage; F, G, I, K-N: Grain filling stage; H, Tilling stage. A-J, Bar=2 mm; K-N, Bar=0.5 mm.
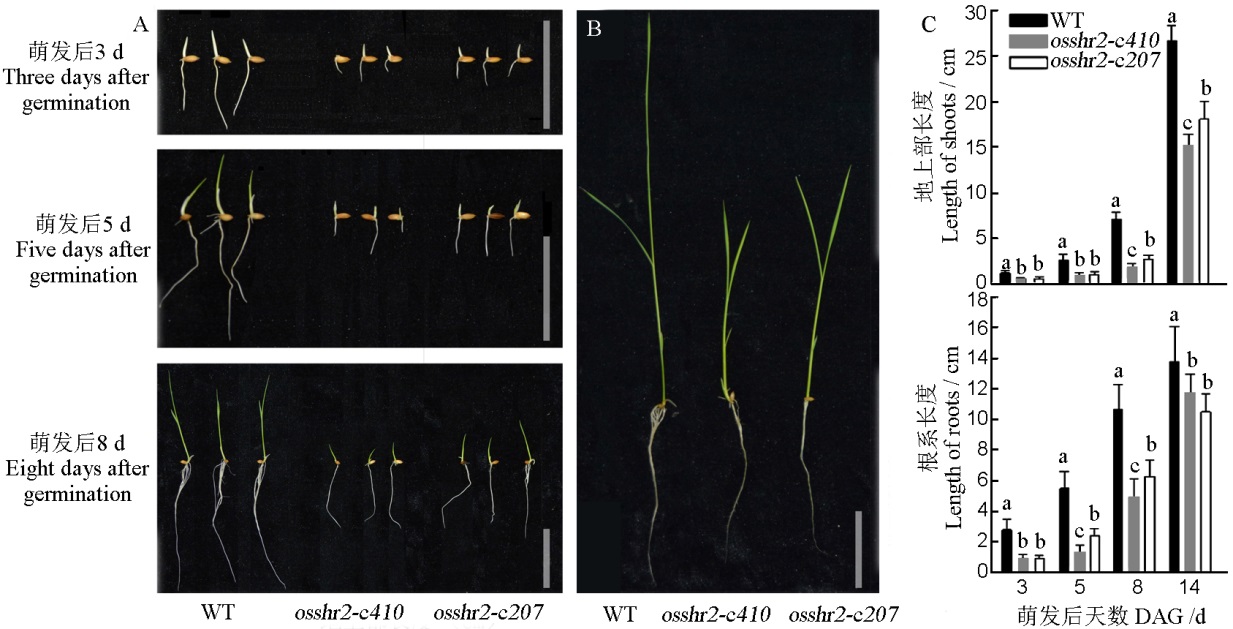
图4 WT和突变体osshr2的萌发及水培下的表型 A–萌发表型(萌发后3 d、5 d和8 d),标尺为5 cm;B–水培表型(萌发后14 d),标尺为5 cm;C–材料地上部和根系长度统计(萌发后3 d、5 d、8 d和14 d);DAG–萌发后天数。
Fig. 4. Phenotype and statistics of seed germination and hydroponics experiments of WT and osshr2. A, Germination phenotype(3, 5 and 8 days after germination), bar=5 cm; B, Hydroponics phenotype(14 days after germination), bar=5 cm; C, Shoot and root length (3, 5, 8 and 14 days after germination); DAG, Day after germination.
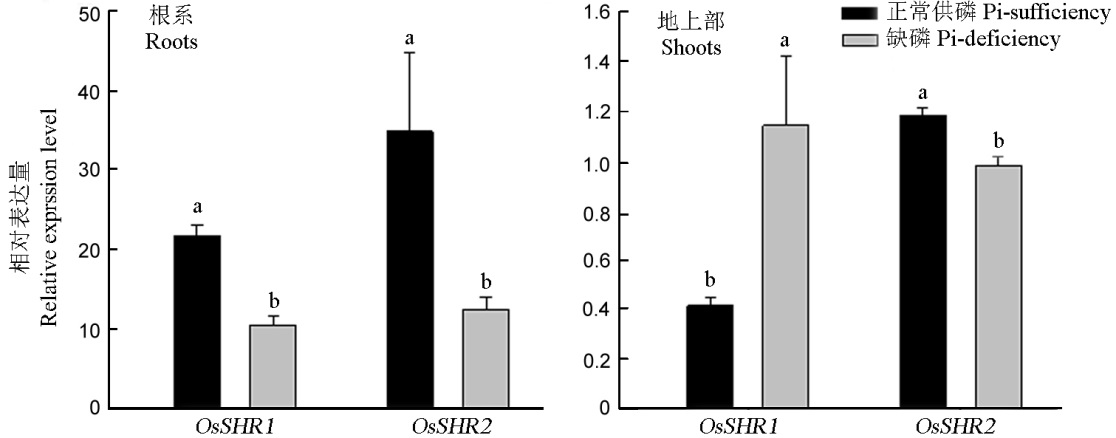
图5 OsSHR1和OsSHR2在水稻正常供磷与缺磷条件下的相对表达量 A–OsSHR1和OsSHR2在水稻地下部的相对表达量;B–OsSHR1和OsSHR2在水稻地上部的相对表达量。
Fig. 5. Relative expression level of OsSHR1 and OsSHR2 under Pi-sufficient and Pi-deficient conditions in rice. A, The relative expression of OsSHR1 and OsSHR2 in the roots; B, The relative expression of OsSHR1 and OsSHR2 in the shoots.
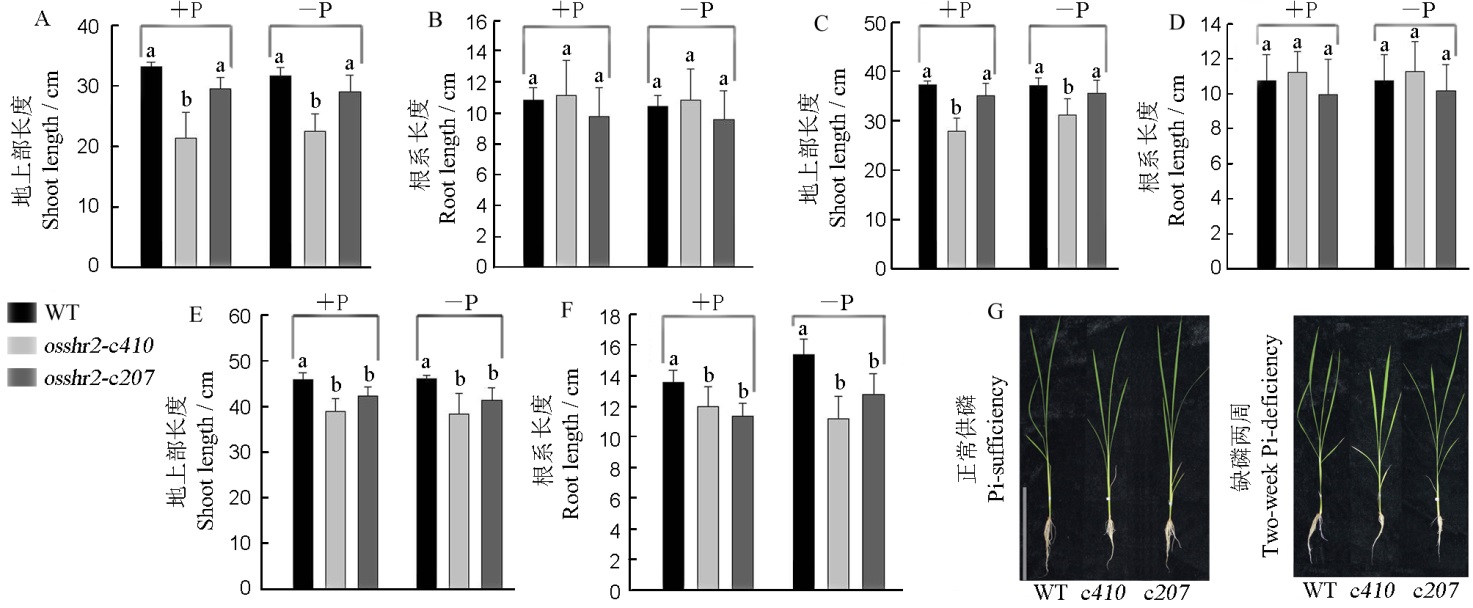
图6 WT和突变体osshr2在正常供磷和缺磷条件下的表型 A和B–缺磷3 d;C和D–缺磷1周;E和F–缺磷2周;G–缺磷2周的表型, 标尺为20 cm。
Fig. 6. Phenotype of the wild type(WT) and osshr2 under Pi-sufficient and Pi-deficient conditions. A and B, Pi-deficient lasting three days; C and D, Pi-deficient lasting one week; E and F, Pi-deficient lasting two weeks; G, The phenotype of material under Pi-sufficient and Pi-deficient lasting two weeks, bar=20 cm. c410, osshr2-c410; c207, osshr2-c207.
| [1] | 朱义旺, 林雅容, 陈亮. 我国水稻分子育种研究进展. 厦门大学学报, 2016, 55(5): 661-671. |
| Zhu Y W, Lin Y R, Chen L.Research progress of rice molecular breeding in China.J Xiamen Univ, 2016, 55(5): 661-671. (in Chinese with English abstract) | |
| [2] | Riechmann J L, Heard J, Martin G, Reuber L, Jiang C Z, Keddie J, Adam L, Pineda O, Ratcliffe O J, Samaha R R, Creelman R, Pilgrim M, Broun P, Zhang J Z, Ghandehari D, Sherman B K, Yu G L.Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science, 2000, 290(5499): 2105-2110. |
| [3] | Pysh L D, Wysocka-Diller J W, Christine C, David B, Benfey P N. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J Cell & Mol Biol, 1999, 18(1): 111. |
| [4] | Cui H, Levesque M P, Vernoux T, Jung J W, Paquette A J, Gallagher K L, Wang J Y, Blilou I, Scheres B, Benfey P N.An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants.Science, 2007, 316(5823): 421-425. |
| [5] | Dolan L.SCARECROWs at the Border.Science, 2007, 316(5823): 377-378. |
| [6] | Wu S, Lee C M, Hayashi T, Pricea S, Divolb F, Henryb S, Pauluzzib G, Perinb C, Gallaghera K L.A plausible mechanism, based upon short-root movement, for regulating the number of cortex cell layers in roots.Proc Natl Acad Sci USA, 2014, 111(45): 16184-16189. |
| [7] | Benfey P N, Scheres B.Root development.Curr Biol, 2000, 10(22): 813-815. |
| [8] | Benfey P N, Linstead P J, Roberts K, Schiefelbein J W, Hauser M T, Aeschbacher R A.Root development in Arabidopsis: Four mutants with dramatically altered root morphogenesis.Development, 1993, 119(1): 57-70. |
| [9] | Laurenzio L D, Wysockadiller J, Malamy J E, Pysh L, Helariutta Y, Freshour G, Hahn M G, Feldmann K A, Benfey P N.The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell, 1996, 86(3): 423-433. |
| [10] | 倪君. OsIAA23介导的生长素信号胚后维持水稻根静止中心. 杭州: 浙江大学, 2011. |
| Ni J.OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in primary in rice. Hangzhou: Zhejiang University, 2011. (in Chinese with English abstract) | |
| [11] | Lim J, Benfey P N.Molecular analysis of the SCARECROW gene in maize reveals a common basis for radial patterning in diverse meristems.Discuss Pap, 2000, 12(8): 1307-1318. |
| [12] | Sbabou L, Bucciarelli B, Miller S, Liu J, Berhada F, Filali-Maltouf A, Allan D, Vance C.Molecular analysis of SCARECROW genes expressed in white lupin cluster roots. J Exp Bot, 2010, 61(5): 1351-1363. |
| [13] | Wang J, Anderssongunneras S, Gaboreanu I, Hertzberg M, Tucker M R, Zheng B, Lesniewska J, Mellerowicz E J, Laux T, Sandberg G, Jones B.Reduced expression of the SHORT-ROOT gene increases the rates of growth and development in hybrid poplar and Arabidopsis. PloS ONE, 2011, 6(12): e28878. |
| [14] | Wysockadiller J W, Helariutta Y, Fukaki H, Malamy J E, Benfey P N.Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development, 2000, 127(3): 595-603. |
| [15] | 霍胜楠. 水稻胚胎发生相关基因的表达及其功能鉴定. 济南: 山东农业大学, 2008. |
| Huo S N.Isolation and characterization of rice genes involved in embryo development. Jinan: Shandong Agricultural University, 2008. (in Chinese with English abstract) | |
| [16] | Cui H, Kong D, Liu X, Hao Y.SCARECROW, SCR-LIKE 23 and SHORT-ROOT control bundle sheath cell fate and function in Arabidopsis thaliana. Plant J Cell & Mol Biol, 2014, 78(2): 319-327. |
| [17] | Gao X R, Wang C L, Cui H C.Identification of bundle sheath cell fate factors provides new tools for C3-to-C4 engineering.Plant Signal & Behav, 2014, 9(6): e29163. |
| [18] | Morikami A.The SCARECROW gene’s role in asymmetric cell divisions in rice plants. Plant J, 2003, 36(1): 45-54. |
| [19] | Lucas M, Swarup R, Paponov I A, Swarup K, Casimiro I, Lake D, Peret B, Zappala S, Mairhofer S, Whitworth M, Wang J H, Ljung K, Marchant A, Sandberg G, Holdsworth M J, Palme K, Pridmore T, Mooney S, Bennett M J.Short-Root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol, 2011, 155(1): 384-398. |
| [20] | Tian H, Jia Y, Niu T, Yu Q, Ding Z.The key players of the primary root growth and development also function in lateral roots in Arabidopsis. Plant Cell Rep, 2014, 33(5): 745-753. |
| [21] | Goh T, Toyokura K, Wells D M, Swarup K, Yamamoto M, Mimura T, Weijers D, Fukaki H, Laplaze L, Bennett M J, Guyomarc’h S.Quiescent center initiation in the Arabidopsis lateral root primordia is dependent on the SCARECROW transcription factor. Development, 1991, 143(18): 3363. |
| [22] | Lavenus J, Goh T, Guyomarc’h S, Hill K, Lucas M, Voß U, Kenobi K, Wilson M H, Farcot E, Hagen G, Guilfoyle T J, Fukaki H, Laplaze L, Bennettb M J.Inference of the Arabidopsis lateral root gene regulatory network suggests a bifurcation mechanism that defines primordia flanking and central zones. Plant Cell, 2015, 27(5): 1368-1388. |
| [23] | Bieleski R.Phosphate pools, phosphate transport, and phosphate availability.Ann Rev Plant Physiol, 1973, 24(1): 225-252. |
| [24] | Muchhal U S, Pardo J M, Raghothama K G.Phosphate transporters from the higher plant Arabidopsis thaliana.Proc Natl Acad Sci USA, 1996, 93(19): 10519-105123. |
| [25] | Wang L, Shan L, Ye Z, Li Z, Du X, Liu D. Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12,AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J Integr Plant Biol, 2014, 56(3): 299-314. |
| [26] | Rausch C, Bucher M.Molecular mechanisms of phosphate transport in plants.Planta, 2002, 216(1): 23-37. |
| [27] | Paszkowski U, Kroken S, Roux C, Briggs S P.Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis.Proc Natl Acad Sci USA, 2002, 99(20): 13324-13329. |
| [28] | Liu F, Chang X J, Ye Y, Xie W B, Wu P, Lian X M.Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice.Mol Plant, 2011, 4(6): 1105-1122. |
| [29] | Zhang F, Sun Y, Pei W, Jain A, Sun R, Cao Y, Wu X N, Jiang T T, Zhang L, Fan X R, Chen A Q, Shen Q R, Xu G H, Sun S B.Involvement of OsPht1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. Plant J Cell & Mol Biol, 2015, 82(4): 556. |
| [30] | Rubio V, Linhares F, Solano R, Martín A C, Iglesias J, Leyva A, Paz-Ares J.A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae.Genes & Dev, 2001, 15(16): 2122-2133. |
| [31] | Bustos R, Castrillo G, Linhares F, Puga M I, Rubio V, Perez-Perez J, Solano R, Leyva A, Paz-Ares J.A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PloS Genet, 2010, 6(9): e1001102. |
| [32] | Wu P, Wang X.Role of OsPHR2 on phosphorus homeostasis and root hairs development in rice(Oryza sativa L.). Plant Signal & Behav, 2008, 3(9): 674-675. |
| [33] | Zhou J, Jiao F, Wu Z C, Li Y Y, Wang X M, He X W, Zhong W Q, Wu P.OsPHR2 is involved in phosphate- starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol, 2008, 146(4): 1673-1686. |
| [34] | Raghothama K G, Maggio A, Narasimhan M L, Kononowicz A K, Wang G, D’Urzo M P, Hasegawa P M, Bressanl R A. Tissue-specific activation of the OsMotin gene by ABA, C2H4 and NaCl involves the same promoter region. Plant Mol Biol, 1997, 34(3): 393-402. |
| [35] | Liao H, Rubio G, Yan X, Cao A, Brown K M, Lynch J P.Effect of phosphorus availability on basal root shallowness in common bean.Plant & Soil, 2001, 232(1): 69-79. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [5] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [6] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [7] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [8] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [9] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [10] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [11] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [12] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [13] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [14] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| [15] | 吕海涛, 李建忠, 鲁艳辉, 徐红星, 郑许松, 吕仲贤. 稻田福寿螺的发生、危害及其防控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 127-139. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||