
中国水稻科学 ›› 2017, Vol. 31 ›› Issue (2): 133-148.DOI: 10.16819/j.1001-7216.2017.6115
傅友强1,2, 于晓莉1, 杨旭健1, 沈宏1,*
出版日期:2017-03-20
发布日期:2017-03-10
通讯作者:
沈宏
基金资助:Youqiang FU1,2, Xiaoli YU1, Xujian YANG1, Hong SHEN1,*
Online:2017-03-20
Published:2017-03-10
Contact:
Hong SHEN
摘要:
【目的】 干湿交替(AWD)是水稻种植过程中最重要的管理方式,能够提高水稻根系活力、增强抗逆性。研究发现,AWD能明显促进水稻根表铁膜的形成。然而,AWD诱导水稻根表铁膜形成的基因表达谱尚未见报道。【方法】 采用砂培试验,研究了长期淹水(CK)、干湿交替(AWD)、长期淹水加Fe2+(CK+Fe)和干湿交替加Fe2+(AWD+Fe)四个处理下水稻根系基因的差异表达谱。【结果】 AWD处理与CK处理相比,水稻根系有506个差异表达基因(DEG)上调表达,有687个DEG下调表达;AWD+Fe处理与CK+Fe处理相比,有308个DEG上调表达和179个DEG下调表达;CK+Fe处理与CK处理相比,有728个DEG上调表达和1175个DEG下调表达;AWD+Fe处理与AWD处理相比,有1252个DEG上调表达和1189个DEG下调表达。维恩图分析发现,共计有3822个DEG参与了AWD诱导水稻根表铁膜形成过程。基因功能(GO)分析表明,在生物过程中共有270个DEG参与了氧化还原反应过程,分子功能中共有165个DEG与氧化还原酶功能有关。生物通路富集分析(KEGG)结果表明,细胞器类、信号刺激类、光合作用类、生物合成类和代谢类等生物通路参与了AWD诱导水稻根表铁膜的形成过程。AWD和根表铁膜形成过程发现有38个共享DEG,这些基因的蛋白注释与水稻抗病性、抗旱性、细胞壁和细胞膜、氧化还原、蛋白激酶和转运、新陈代谢过程有关。与CK处理相比,AWD处理诱导了氧化还原反应过程相关的基因达102个,占总DEG数的8.25%。AWD处理促进了水稻根系过氧化物酶、脂肪酸氧化酶和乙醇酸氧化酶等相关基因的上调表达。与CK处理相比,AWD处理提高了水稻根系活力和根表铁膜数量,分别为22.9%和45.7%。实时荧光定量PCR验证结果与转录组分析结果基本一致,其相关系数达到0.90。【结论】综上所述,AWD明显诱导了氧化还原反应过程中的相关基因大量表达,增加了根系氧化力,促进根表铁膜的形成;过氧化物酶、脂肪酸氧化酶和乙醇酸氧化酶等相关的基因可能是AWD诱导水稻根表铁膜形成的重要基因。
傅友强, 于晓莉, 杨旭健, 沈宏. 干湿交替诱导水稻根表铁膜形成的基因表达谱分析[J]. 中国水稻科学, 2017, 31(2): 133-148.
Youqiang FU, Xiaoli YU, Xujian YANG, Hong SHEN. Gene Expression Profile Analysis for Alternate Wetting and Drying Induced Formation of Iron Plaque on Root Surface of Rice Seedlings[J]. Chinese Journal OF Rice Science, 2017, 31(2): 133-148.
| 基因 Gene | 荧光定量PCR正反引物 Forward and reverse primers of qRT-PCR | Tm | GC/% | 产物长度 Product length/bp | |
|---|---|---|---|---|---|
| BGIOSGA001629-TA | F: 5’-CGCCGTCTTCTTCTGCCAGAT-3’ R: 5’-GGTCACTTGCTTGTCCGTGCTT-3’ | 59.3 | 55.8 | 157 | |
| BGIOSGA026866-TA | F: 5’-TTCTACCTGGTGGCGCTGCT-3’ R: 5’-TCGACGAAGCGGCTCCACTT-3’ | 59.7 | 60.0 | 200 | |
| BGIOSGA029195-TA | F: 5’-GAGGTACGGCACCAGCTTCAAC-3’ R: 5’- CCGCCTCCATCACCTCCATGAA-3’ | 60.1 | 59.1 | 188 | |
| BGIOSGA014041-TA | F: 5’-GCCTCCTCCGCATCTTCTTCCA-3’ R: 5’-CGTGGTTGCAGAGTCAGGTTGG-3’ | 60.2 | 59.1 | 115 | |
| BGIOSGA014859-TA | F: 5’-TACTACACGGCGGAGCAGGAGA-3’ R: 5’-TTGCAGCTCTTCTTGGCGGACT-3’ | 60.8 | 56.8 | 173 | |
| BGIOSGA030543-TA | F: 5’-GGGCTCATCGGGTTCATCTGGA-3’ R: 5’-GCAGGCAATGTGGTGGGTGTT-3’ | 59.9 | 58.3 | 177 | |
| BGIOSGA033475-TA | F: 5’-GAGTGGCAGCAGCAGCATTAGT-3’ R: 5’-ACCTGTGTACGGATCGCCTCTC-3’ | 59.7 | 56.8 | 107 | |
| BGIOSGA009910-TA | F: 5’-GCTTGGTCGCCGTGTTGGAA-3’ R: 5’-GCATCGCCTCTTTCTCGCTGAA-3’ | 59.5 | 57.3 | 117 | |
| BGIOSGA017964-TA | F: 5’-TGGAGCAAGGCGGAGGACAA-3’ R: 5’-CCACGAGCACCTGGTAGTGTTC-3’ | 59.3 | 59.5 | 142 | |
| OsActin | F: 5’-CCGCCAGAGAGGAAGTACAG-3’ R: 5’-AAGCACTTCCTGTGGACGAT-3’ | 59.4 | 55.1 | 131 | |
表1 实时荧光定量PCR引物
Table 1 Primers of quantitative real-time PCR.
| 基因 Gene | 荧光定量PCR正反引物 Forward and reverse primers of qRT-PCR | Tm | GC/% | 产物长度 Product length/bp | |
|---|---|---|---|---|---|
| BGIOSGA001629-TA | F: 5’-CGCCGTCTTCTTCTGCCAGAT-3’ R: 5’-GGTCACTTGCTTGTCCGTGCTT-3’ | 59.3 | 55.8 | 157 | |
| BGIOSGA026866-TA | F: 5’-TTCTACCTGGTGGCGCTGCT-3’ R: 5’-TCGACGAAGCGGCTCCACTT-3’ | 59.7 | 60.0 | 200 | |
| BGIOSGA029195-TA | F: 5’-GAGGTACGGCACCAGCTTCAAC-3’ R: 5’- CCGCCTCCATCACCTCCATGAA-3’ | 60.1 | 59.1 | 188 | |
| BGIOSGA014041-TA | F: 5’-GCCTCCTCCGCATCTTCTTCCA-3’ R: 5’-CGTGGTTGCAGAGTCAGGTTGG-3’ | 60.2 | 59.1 | 115 | |
| BGIOSGA014859-TA | F: 5’-TACTACACGGCGGAGCAGGAGA-3’ R: 5’-TTGCAGCTCTTCTTGGCGGACT-3’ | 60.8 | 56.8 | 173 | |
| BGIOSGA030543-TA | F: 5’-GGGCTCATCGGGTTCATCTGGA-3’ R: 5’-GCAGGCAATGTGGTGGGTGTT-3’ | 59.9 | 58.3 | 177 | |
| BGIOSGA033475-TA | F: 5’-GAGTGGCAGCAGCAGCATTAGT-3’ R: 5’-ACCTGTGTACGGATCGCCTCTC-3’ | 59.7 | 56.8 | 107 | |
| BGIOSGA009910-TA | F: 5’-GCTTGGTCGCCGTGTTGGAA-3’ R: 5’-GCATCGCCTCTTTCTCGCTGAA-3’ | 59.5 | 57.3 | 117 | |
| BGIOSGA017964-TA | F: 5’-TGGAGCAAGGCGGAGGACAA-3’ R: 5’-CCACGAGCACCTGGTAGTGTTC-3’ | 59.3 | 59.5 | 142 | |
| OsActin | F: 5’-CCGCCAGAGAGGAAGTACAG-3’ R: 5’-AAGCACTTCCTGTGGACGAT-3’ | 59.4 | 55.1 | 131 | |
| 处理 Treatment | 总原始读序数Total raw reads | 接头污染 Adaptor pollution | 多N序列 More N sequences | 低质序列 Low sequences | 干净序列 Clear sequences | Clean-Q20/% |
|---|---|---|---|---|---|---|
| CK-1 | 15 203 921 (100%) | 7 879 (0.05%) | 8 096 (0.05%) | 226 110 (1.49%) | 14 961 836 (98.41%) | 99.03 |
| CK-2 | 14 617 585 (100%) | 3 596 (0.02%) | 7 679 (0.05%) | 218 591 (1.50%) | 14 387 719 (98.43%) | 99.02 |
| CK-3 | 16 294 296 (100%) | 4 603 (0.03%) | 8 909 (0.05%) | 239 703 (1.47%) | 16 041 081 (98.45%) | 99.03 |
| AWD-1 | 15 761 265 (100%) | 7 966 (0.05%) | 8 422 (0.05%) | 243 872 (1.55%) | 15 501 005 (98.35%) | 98.99 |
| AWD-2 | 17 182 802 (100%) | 6 061 (0.04%) | 9 178 (0.05%) | 261 475 (1.52%) | 16 906 088 (98.39%) | 99.00 |
| AWD-3 | 15 568 129 (100%) | 9 138 (0.06%) | 8 057 (0.05%) | 235 384 (1.51%) | 15 315 550 (98.38%) | 99.02 |
| CK+Fe-1 | 14 026 907 (100%) | 7 217 (0.05%) | 7 497 (0.05%) | 210 847 (1.50%) | 13 801 346 (98.39%) | 99.01 |
| CK+Fe-2 | 15 543 510 (100%) | 13 312 (0.09%) | 8 186 (0.05%) | 225 777 (1.45%) | 15 296 235 (98.41%) | 99.04 |
| CK+Fe-3 | 17 756 771 (100%) | 45 874 (0.26%) | 576 (0.00%) | 271 929 (1.53%) | 17 438 392 (98.21%) | 98.77 |
| AWD+Fe-1 | 16 562 694 (100%) | 25 236 (0.15%) | 487 (0.00%) | 276 876 (1.67%) | 16 260 095 (98.17%) | 98.65 |
| AWD+Fe-2 | 17 369 964 (100%) | 39 004 (0.22%) | 544 (0.00%) | 265 196 (1.53%) | 17 065 220 (98.25%) | 98.78 |
| AWD+Fe-3 | 13 292 393 (100%) | 11 353 (0.09%) | 1 501 (0.01%) | 197 158 (1.48%) | 13 082 381 (98.42%) | 98.97 |
表2 数据过滤统计分析
Table 2 Statistical analysis of data filtering.
| 处理 Treatment | 总原始读序数Total raw reads | 接头污染 Adaptor pollution | 多N序列 More N sequences | 低质序列 Low sequences | 干净序列 Clear sequences | Clean-Q20/% |
|---|---|---|---|---|---|---|
| CK-1 | 15 203 921 (100%) | 7 879 (0.05%) | 8 096 (0.05%) | 226 110 (1.49%) | 14 961 836 (98.41%) | 99.03 |
| CK-2 | 14 617 585 (100%) | 3 596 (0.02%) | 7 679 (0.05%) | 218 591 (1.50%) | 14 387 719 (98.43%) | 99.02 |
| CK-3 | 16 294 296 (100%) | 4 603 (0.03%) | 8 909 (0.05%) | 239 703 (1.47%) | 16 041 081 (98.45%) | 99.03 |
| AWD-1 | 15 761 265 (100%) | 7 966 (0.05%) | 8 422 (0.05%) | 243 872 (1.55%) | 15 501 005 (98.35%) | 98.99 |
| AWD-2 | 17 182 802 (100%) | 6 061 (0.04%) | 9 178 (0.05%) | 261 475 (1.52%) | 16 906 088 (98.39%) | 99.00 |
| AWD-3 | 15 568 129 (100%) | 9 138 (0.06%) | 8 057 (0.05%) | 235 384 (1.51%) | 15 315 550 (98.38%) | 99.02 |
| CK+Fe-1 | 14 026 907 (100%) | 7 217 (0.05%) | 7 497 (0.05%) | 210 847 (1.50%) | 13 801 346 (98.39%) | 99.01 |
| CK+Fe-2 | 15 543 510 (100%) | 13 312 (0.09%) | 8 186 (0.05%) | 225 777 (1.45%) | 15 296 235 (98.41%) | 99.04 |
| CK+Fe-3 | 17 756 771 (100%) | 45 874 (0.26%) | 576 (0.00%) | 271 929 (1.53%) | 17 438 392 (98.21%) | 98.77 |
| AWD+Fe-1 | 16 562 694 (100%) | 25 236 (0.15%) | 487 (0.00%) | 276 876 (1.67%) | 16 260 095 (98.17%) | 98.65 |
| AWD+Fe-2 | 17 369 964 (100%) | 39 004 (0.22%) | 544 (0.00%) | 265 196 (1.53%) | 17 065 220 (98.25%) | 98.78 |
| AWD+Fe-3 | 13 292 393 (100%) | 11 353 (0.09%) | 1 501 (0.01%) | 197 158 (1.48%) | 13 082 381 (98.42%) | 98.97 |
| 处理 Treatment | 有效Reads数Effective reads | 总比对数 Total mapped | 多序列比对Multiple mapped | 唯一比对Uniquely mapped | 正链比对 Reads map to '+' | 负链比对Reads map to '-' |
|---|---|---|---|---|---|---|
| CK-1 | 14 961 836 (100%) | 14 064 974 (94.01%) | 1 399 518 (9.35%) | 12 665 456 (84.65%) | 6 682 511 (44.66%) | 7 382 463 (49.34%) |
| CK-2 | 14 387 719 (100%) | 13 418 012 (93.26%) | 1 304 324 (9.07%) | 12 113 688 (84.19%) | 6 395 522 (44.45%) | 7 022 490 (48.81%) |
| CK-3 | 16 041 081 (100%) | 15 096 703 (94.11%) | 1 252 800 (7.81%) | 13 843 903 (86.30%) | 6 853 649 (44.75%) | 7 814 056 (48.71%) |
| AWD-1 | 15 501 005 (100%) | 14 458 761 (93.28%) | 1 408 866 (9.09%) | 13 049 895 (84.19%) | 6 921 799 (44.65%) | 7 536 962 (48.62%) |
| AWD-2 | 16 906 088 (100%) | 15 898 780 (94.04%) | 2 003 341 (11.85%) | 13 895 439 (82.19%) | 7 409 954 (43.83%) | 8 488 826 (50.21%) |
| AWD-3 | 15 315 550 (100%) | 14 292 666 (93.32%) | 1 286 870 (8.40%) | 13 005 796 (84.92%) | 6 853 649 (44.75%) | 7 439 017 (48.57%) |
| CK+Fe-1 | 13 801 346 (100%) | 12 827 342 (92.94%) | 1 270 100 (9.20%) | 11 557 242 (83.74%) | 6 111 387 (44.28%) | 6 715 955 (48.66%) |
| CK+Fe-2 | 15 296 235 (100%) | 14 278 682 (93.35%) | 1 565 420 (10.23%) | 12 713 262 (83.11%) | 6 757 606 (44.18%) | 7 521 076 (49.17%) |
| CK+Fe-3 | 17 438 392 (100%) | 16 001 385 (91.76%) | 1 368 718 (7.85%) | 14 632 667 (83.91%) | 7 756 673 (44.48%) | 8 244 712 (47.28%) |
| AWD+Fe-1 | 16 260 095 (100%) | 15 235 534 (93.70%) | 1 559 671 (9.59%) | 13 675 863 (84.11%) | 7 277 032 (44.75%) | 7 958 502 (48.94%) |
| AWD+Fe-2 | 17 065 220 (100%) | 15 859 868 (92.94%) | 1 430 141 (8.38%) | 14 429 727 (84.56%) | 7 619 349 (44.65%) | 8 240 519 (48.29%) |
| AWD+Fe-3 | 13 082 381 (100%) | 12 121 909 (92.66%) | 1 093 176 (8.36%) | 11 028 733 (84.30%) | 5 830 048 (44.56%) | 6 291 861 (48.09%) |
表 3 Reads比对结果统计
Table 3 Alignment result statistics of Reads.
| 处理 Treatment | 有效Reads数Effective reads | 总比对数 Total mapped | 多序列比对Multiple mapped | 唯一比对Uniquely mapped | 正链比对 Reads map to '+' | 负链比对Reads map to '-' |
|---|---|---|---|---|---|---|
| CK-1 | 14 961 836 (100%) | 14 064 974 (94.01%) | 1 399 518 (9.35%) | 12 665 456 (84.65%) | 6 682 511 (44.66%) | 7 382 463 (49.34%) |
| CK-2 | 14 387 719 (100%) | 13 418 012 (93.26%) | 1 304 324 (9.07%) | 12 113 688 (84.19%) | 6 395 522 (44.45%) | 7 022 490 (48.81%) |
| CK-3 | 16 041 081 (100%) | 15 096 703 (94.11%) | 1 252 800 (7.81%) | 13 843 903 (86.30%) | 6 853 649 (44.75%) | 7 814 056 (48.71%) |
| AWD-1 | 15 501 005 (100%) | 14 458 761 (93.28%) | 1 408 866 (9.09%) | 13 049 895 (84.19%) | 6 921 799 (44.65%) | 7 536 962 (48.62%) |
| AWD-2 | 16 906 088 (100%) | 15 898 780 (94.04%) | 2 003 341 (11.85%) | 13 895 439 (82.19%) | 7 409 954 (43.83%) | 8 488 826 (50.21%) |
| AWD-3 | 15 315 550 (100%) | 14 292 666 (93.32%) | 1 286 870 (8.40%) | 13 005 796 (84.92%) | 6 853 649 (44.75%) | 7 439 017 (48.57%) |
| CK+Fe-1 | 13 801 346 (100%) | 12 827 342 (92.94%) | 1 270 100 (9.20%) | 11 557 242 (83.74%) | 6 111 387 (44.28%) | 6 715 955 (48.66%) |
| CK+Fe-2 | 15 296 235 (100%) | 14 278 682 (93.35%) | 1 565 420 (10.23%) | 12 713 262 (83.11%) | 6 757 606 (44.18%) | 7 521 076 (49.17%) |
| CK+Fe-3 | 17 438 392 (100%) | 16 001 385 (91.76%) | 1 368 718 (7.85%) | 14 632 667 (83.91%) | 7 756 673 (44.48%) | 8 244 712 (47.28%) |
| AWD+Fe-1 | 16 260 095 (100%) | 15 235 534 (93.70%) | 1 559 671 (9.59%) | 13 675 863 (84.11%) | 7 277 032 (44.75%) | 7 958 502 (48.94%) |
| AWD+Fe-2 | 17 065 220 (100%) | 15 859 868 (92.94%) | 1 430 141 (8.38%) | 14 429 727 (84.56%) | 7 619 349 (44.65%) | 8 240 519 (48.29%) |
| AWD+Fe-3 | 13 082 381 (100%) | 12 121 909 (92.66%) | 1 093 176 (8.36%) | 11 028 733 (84.30%) | 5 830 048 (44.56%) | 6 291 861 (48.09%) |
| 基因Gene | Log2(AWD/CK) | Log2(AWD+Fe/CK) | 非冗余蛋白质序列数据库蛋白注释NR-annotation | ||
|---|---|---|---|---|---|
| 抗病性类Disease resistance categories | |||||
| BGIOSGA033475-TA | 4.47 | 6.45 | Pathogen-related protein Rir1a | ||
| BGIOSGA032127-TA | -1.95 | 0.00 | Plant disease resistance response protein family | ||
| 抗旱性类 Drought resistance categories | |||||
| BGIOSGA029417-TA | -1.22 | 0.73 | Dehydration-responsive element-binding protein 1B | ||
| BGIOSGA011664-TA | -1.36 | 0.35 | Hydrophobic Protein from Soybean (HPS)-like | ||
| BGIOSGA016950-TA | -1.42 | 0.54 | Dehydration-responsive element-binding protein 1E | ||
| BGIOSGA008804-TA | -1.67 | 0.29 | Dehydration-responsive element-binding protein 1G | ||
| 细胞壁和细胞膜类Cell wall and cell membrane categories | |||||
| BGIOSGA005143-TA | 2.85 | 4.02 | Beta-1,3-glucanase precursor | ||
| BGIOSGA012450-TA | -1.17 | 0.00 | Cotton fibre expressed protein | ||
| BGIOSGA033296-TA | -1.19 | 0.09 | Putative membrane protein | ||
| BGIOSGA012438-TA | -1.36 | 0.00 | Cellulose microfibril orientation-like protein | ||
| BGIOSGA009910-TA | -1.72 | -2.96 | Microtubule associated protein | ||
| BGIOSGA025169-TA | -2.33 | -1.11 | Retinal pigment epithelial membrane protein | ||
| BGIOSGA019479-TA | -4.69 | 1.01 | Glycosyl hydrolases family 18 | ||
| BGIOSGA031806-TA | -5.10 | 2.20 | Lycine-rich cell wall structural protein 2 precursor | ||
| 氧化还原类Redox categories | |||||
| BGIOSGA017163-TA | 1.15 | -0.92 | Peroxisomal (S)-2-hydroxy-acid oxidase GLO3 | ||
| BGIOSGA005230-TA | -1.31 | -4.49 | Peroxidase | ||
| BGIOSGA008063-TA | -1.47 | -4.24 | Ubiquinol oxidase | ||
| BGIOSGA002551-TA | -1.50 | 0.71 | Isoflavone reductase-like protein | ||
| BGIOSGA026226-TA | -3.74 | -5.10 | 1-Cys peroxiredoxin B | ||
| 蛋白激酶和转运类 Protein kinase and transfer categories | |||||
| BGIOSGA000958-TA | -1.46 | -0.37 | Protein kinase-like domain containing protein | ||
| BGIOSGA017285-TA | -1.55 | 0.74 | Cysteine-rich receptor-like protein kinase 10-like | ||
| BGIOSGA025234-TA | -1.74 | -3.39 | Plant lipid transfer protein | ||
| BGIOSGA003951-TA | -2.63 | -0.73 | Bidirectional sugar transporter SWEET6a | ||
| BGIOSGA010483-TA | -2.67 | -0.52 | Cytosolic pyruvate orthophosphate dikinase | ||
| 新陈代谢类Metabolism categories | |||||
| BGIOSGA012186-TA | 2.67 | 1.61 | Non-symbiotic hemoglobin 2-like | ||
| BGIOSGA003133-TA | 1.94 | 4.95 | Light-harvesting-like protein 3; Provisional | ||
| BGIOSGA015910-TA | 1.17 | 3.07 | Peptidase C1A subfamily | ||
| BGIOSGA026338-TA | -1.03 | -3.21 | Rossmann-fold NAD(P)(+)-binding proteins | ||
| BGIOSGA033178-TA | -1.10 | -0.10 | Late embryogenesis abundant protein | ||
| BGIOSGA031475-TA | -1.41 | -2.88 | Glutathione S-transferase GST 11 | ||
| BGIOSGA017447-TA | -1.48 | -0.25 | Peptidase C13 family | ||
| BGIOSGA023256-TA | -1.74 | -0.24 | Chloroplast Nucleoids DNA-binding Protease | ||
| BGIOSGA016118-TA | -1.76 | -0.07 | Peptidase C1A subfamily | ||
| BGIOSGA010778-TA | -2.16 | -0.33 | Alanine: glyoxylate aminotransferase-like | ||
| 其他 Others | |||||
| NCRNA_4800 | 2.33 | -0.10 | No protein | ||
| BGIOSGA033067-TA | 2.15 | 0.89 | Hypothetical protein | ||
| BGIOSGA031784-TA | 2.08 | -0.18 | Conserved hypothetical protein | ||
| BGIOSGA013300-TA | -1.86 | -3.72 | Hypothetical protein | ||
表4 38个差异表达基因的分类及注释
Table 4 Categories and annotations of 38 differentially expressed genes.
| 基因Gene | Log2(AWD/CK) | Log2(AWD+Fe/CK) | 非冗余蛋白质序列数据库蛋白注释NR-annotation | ||
|---|---|---|---|---|---|
| 抗病性类Disease resistance categories | |||||
| BGIOSGA033475-TA | 4.47 | 6.45 | Pathogen-related protein Rir1a | ||
| BGIOSGA032127-TA | -1.95 | 0.00 | Plant disease resistance response protein family | ||
| 抗旱性类 Drought resistance categories | |||||
| BGIOSGA029417-TA | -1.22 | 0.73 | Dehydration-responsive element-binding protein 1B | ||
| BGIOSGA011664-TA | -1.36 | 0.35 | Hydrophobic Protein from Soybean (HPS)-like | ||
| BGIOSGA016950-TA | -1.42 | 0.54 | Dehydration-responsive element-binding protein 1E | ||
| BGIOSGA008804-TA | -1.67 | 0.29 | Dehydration-responsive element-binding protein 1G | ||
| 细胞壁和细胞膜类Cell wall and cell membrane categories | |||||
| BGIOSGA005143-TA | 2.85 | 4.02 | Beta-1,3-glucanase precursor | ||
| BGIOSGA012450-TA | -1.17 | 0.00 | Cotton fibre expressed protein | ||
| BGIOSGA033296-TA | -1.19 | 0.09 | Putative membrane protein | ||
| BGIOSGA012438-TA | -1.36 | 0.00 | Cellulose microfibril orientation-like protein | ||
| BGIOSGA009910-TA | -1.72 | -2.96 | Microtubule associated protein | ||
| BGIOSGA025169-TA | -2.33 | -1.11 | Retinal pigment epithelial membrane protein | ||
| BGIOSGA019479-TA | -4.69 | 1.01 | Glycosyl hydrolases family 18 | ||
| BGIOSGA031806-TA | -5.10 | 2.20 | Lycine-rich cell wall structural protein 2 precursor | ||
| 氧化还原类Redox categories | |||||
| BGIOSGA017163-TA | 1.15 | -0.92 | Peroxisomal (S)-2-hydroxy-acid oxidase GLO3 | ||
| BGIOSGA005230-TA | -1.31 | -4.49 | Peroxidase | ||
| BGIOSGA008063-TA | -1.47 | -4.24 | Ubiquinol oxidase | ||
| BGIOSGA002551-TA | -1.50 | 0.71 | Isoflavone reductase-like protein | ||
| BGIOSGA026226-TA | -3.74 | -5.10 | 1-Cys peroxiredoxin B | ||
| 蛋白激酶和转运类 Protein kinase and transfer categories | |||||
| BGIOSGA000958-TA | -1.46 | -0.37 | Protein kinase-like domain containing protein | ||
| BGIOSGA017285-TA | -1.55 | 0.74 | Cysteine-rich receptor-like protein kinase 10-like | ||
| BGIOSGA025234-TA | -1.74 | -3.39 | Plant lipid transfer protein | ||
| BGIOSGA003951-TA | -2.63 | -0.73 | Bidirectional sugar transporter SWEET6a | ||
| BGIOSGA010483-TA | -2.67 | -0.52 | Cytosolic pyruvate orthophosphate dikinase | ||
| 新陈代谢类Metabolism categories | |||||
| BGIOSGA012186-TA | 2.67 | 1.61 | Non-symbiotic hemoglobin 2-like | ||
| BGIOSGA003133-TA | 1.94 | 4.95 | Light-harvesting-like protein 3; Provisional | ||
| BGIOSGA015910-TA | 1.17 | 3.07 | Peptidase C1A subfamily | ||
| BGIOSGA026338-TA | -1.03 | -3.21 | Rossmann-fold NAD(P)(+)-binding proteins | ||
| BGIOSGA033178-TA | -1.10 | -0.10 | Late embryogenesis abundant protein | ||
| BGIOSGA031475-TA | -1.41 | -2.88 | Glutathione S-transferase GST 11 | ||
| BGIOSGA017447-TA | -1.48 | -0.25 | Peptidase C13 family | ||
| BGIOSGA023256-TA | -1.74 | -0.24 | Chloroplast Nucleoids DNA-binding Protease | ||
| BGIOSGA016118-TA | -1.76 | -0.07 | Peptidase C1A subfamily | ||
| BGIOSGA010778-TA | -2.16 | -0.33 | Alanine: glyoxylate aminotransferase-like | ||
| 其他 Others | |||||
| NCRNA_4800 | 2.33 | -0.10 | No protein | ||
| BGIOSGA033067-TA | 2.15 | 0.89 | Hypothetical protein | ||
| BGIOSGA031784-TA | 2.08 | -0.18 | Conserved hypothetical protein | ||
| BGIOSGA013300-TA | -1.86 | -3.72 | Hypothetical protein | ||
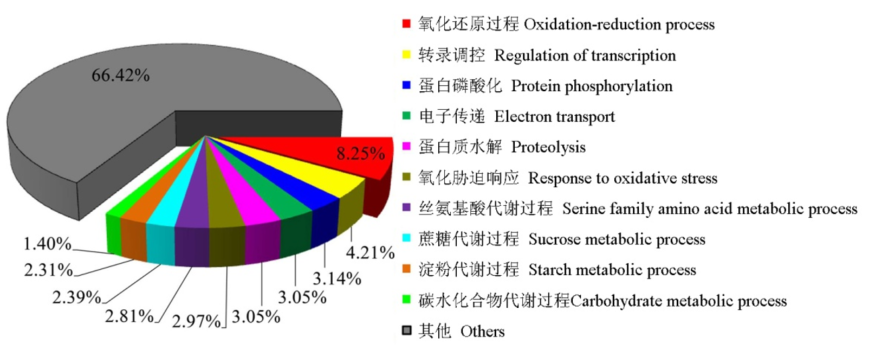
图6 AWD vs CK数据库中的差异表达基因在生物过程(GO)中的分布情况
Fig. 6. Distribution of differentially expressed genes in the biological process of GO classification in AWD vs CK library.
| [1] | Xiao W D, Ye X Z, Yang X E, Li T Q, Zhao S P, Zhang Q.Effects of alternating wetting and drying versus continuous flooding on chromium fate in paddy soils.Ecotox Environ Safe, 2015, 113: 439-445. |
| [2] | Ye Y S, Liang X Q, Chen Y X, Liu J, Gu J T, Guo R, Li L.Alternate wetting and drying irrigation and controlled-release nitrogen fertilizer in late-season rice. Effects on dry matter accumulation, yield, water and nitrogen use.Field Crop Res, 2013, 144: 212-224. |
| [3] | 刘艳,孙文涛,宫亮,蔡广兴. 水分调控对水稻根际土壤及产量的影响. 灌溉排水学报, 2014, 33(2): 98-100. |
| Liu Y, Sun W T, Gong L, Cai G X.Effects of water regulation of rhizosphere soil and yield of rice.J Irrig Drain, 2014, 33(2): 98-100. (in Chinese with English abstract) | |
| [4] | 丁汉卿, 赖聪玲, 沈宏. 干湿交替和过氧化物对水稻根表铁膜及养分吸收的影响. 生态环境学报, 2015, 24(12): 1983-1988. |
| Ding H Q, Lai C L, Shen H.Influence of alternative wetting and drying and peroxides on the formation of iron plaque on rice root surface and nutrient uptake.Ecol Environ Sci, 2015, 24(12): 1983-1988. (in Chinese with English abstract) | |
| [5] | Pandey P, Srivastava R K, Rajpoot R, Rani A, Pandey A K, Dubey R S.Water deficit and aluminum interactive effects on generation of reactive oxygen species and responses of antioxidative enzymes in the seedlings of two rice cultivars differing in stress tolerance.Environ Sci Pollut Res, 2016, 23(2): 1516-1528. |
| [6] | 蔡昆争,吴学祝,骆世明,王维. 不同生育期水分胁迫对水稻根系活力、叶片水势和保护酶活性的影响. 华南农业大学学报, 2008, 29(2): 7-10. |
| Cai K Z, Wu X Z, Luo S M, Wang W.Effects of water stress at different growth stages on root activity, leaf water potential and protective enzymes activity in rice.J South China Agric Univ, 2008, 29(2): 7-10. (in Chinese with English abstract) | |
| [7] | Wang X L, Wang J J, Sun R H, Hou X G, Zhao W, Shi J, Zhang Y F, Qi L, Li X L, Dong P H, Zhang L X, Xu G W, Gan H B.Correlation of the corn compensatory growth mechanism after post-drought rewatering with cytokinin induced by root nitrate absorption.Agric Water Manag, 2016, 166: 77-85. |
| [8] | 傅友强,于智卫,蔡昆争,沈宏. 水稻根表铁膜形成机制及其生态环境效应. 植物营养与肥料学报, 2010, 16(6): 1527-1534. |
| Fu Y Q, Yu Z W, Cai K Z, Shen H.Mechanisms of iron plaque formation on root surface of rice plants and their ecological and environmental effects: A review.Plant Nutr Fert Sci, 2010, 16(6): 1527-1534. (in Chinese with English abstract) | |
| [9] | Fu Y Q, Yang X J, Ye Z H, Shen H.Identification, separation and component analysis of reddish brown and non-reddish brown iron plaque on rice (Oryza sativa) root surface. Plant Soil, 2016, 402(1): 277-290. |
| [10] | 钟顺清. 根表铁膜对2种景观湿地植物根系发育及活力的影响. 水生态学杂志, 2015, 36(1): 74-79. |
| Zhong S Q.Effect of iron plaque on root growth and activity of two wetland plants.J Hydroecol, 2015, 36(1): 74-79. (in Chinese with English abstract) | |
| [11] | Pandey A, Sharma M, Pandey G.Elucidation of abiotic stress signaling in plants. New York: Springer, 2015: 231-270. |
| [12] | Sun J, Xie D W, Zhao H W, Zou D T.Genome-wide identification of the class III aminotransferase gene family in rice and expression analysis under abiotic stress.Genes Genom, 2013, 35(5): 597-608. |
| [13] | Huda K M K, Yadav S, Banu M S A, Trivedi D K, Tuteja N. Genome-wide analysis of plant-type II Ca2+ ATPases gene family from rice and arabidopsis: Potential role in abiotic stresses.Plant Physiol Bioch, 2013, 65: 32-47. |
| [14] | Singh A, Giri J, Kapoor S, Tyagi A K, Pandey G K.Protein phosphatase complement in rice: Genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development.BMC Genom, 2010, 11(435). |
| [15] | Ray S, Agarwal P, Arora R, Kapoor S, Tyagi A K.Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp indica). Mol Genet Genom, 2007, 278(5): 493-505. |
| [16] | Venu R C, Sreerekha M V, Madhav M S, Nobuta K, Mohan K M, Chen S B, Jia Y L, Meyers B C, Wang G L.Deep transcriptome sequencing reveals the expression of key functional and regulatory genes involved in the abiotic stress signaling pathways in rice.J Plant Biol, 2013, 56(4): 216-231. |
| [17] | Jiang S Y, Ma A, Ramamoorthy R, Ramachandran S.Genome-wide survey on genomic variation, expression divergence, and evolution in two contrasting rice genotypes under high salinity stress.Genom Biol Evol, 2013, 5(11): 2032-2050. |
| [18] | Mal C, Deb A, Aftabuddin M, Kundu S.A network analysis of miRNA mediated gene regulation of rice: Crosstalk among biological processes.Mol Biosyst, 2015, 11(8): 2273-2280. |
| [19] | Jangam A P, Pathak R R, Raghuram N.Microarray analysis of rice d1 (RGA1) mutant reveals the potential role of G-protein alpha subunit in regulating multiple abiotic stresses such as drought, salinity, heat, and cold. Front Plant Sci, 2016, 7: 11. |
| [20] | Yoshida S, Douglas A, Forno J.Laboratory manual for physiological studies of rice. Philippines: International Rice Research Institute, 1976: 57-65. |
| [21] | 傅友强, 杨旭健, 吴道铭, 沈宏. 磷素对水稻根表红棕色铁膜的影响及营养效应. 中国农业科学, 2014, 47(6): 1072-1085. |
| Fu Y Q, Yang X J, Wu D M, Shen H.Effect of phosphorus on reddish brown iron plaque on root surface of rice seedlings and their nutritional effects.Agric Sci China, 2014, 47(6): 1072-1085.(in Chinese with English abstract) | |
| [22] | Pathak R R, Lochab S.A method for rapid isolation of total RNA of high purity and yield fromArthrospira platensis. Can J Microbiol, 2010, 56(7): 578-584. |
| [23] | Taylor G, Crowder A.Use of the DCB technique for extraction of hydrous iron oxides from roots of wetland plant.Am J Bot, 1983, 70: 1254-1257. |
| [24] | Ramasamy S H. Berge T, Purushothaman S.Yield formation in rice in response to drainage and nitrogen application.Field Crop Res, 1997, 51: 65-82. |
| [25] | Imadi S R, Kazi A G, Ahanger M A, Gucel S, Ahmad P.Plant transcriptomics and responses to environmental stress: An overview.J Genet, 2015, 94(3): 525-537. |
| [26] | Neto J B, Frei M.Microarray Meta-analysis focused on the response of genes involved in redox homeostasis to diverse abiotic stresses in rice.Front Plant Sci, 2016, 6: 1260. |
| [27] | Tian X J, Long Y, Wang J, Zhang J W, Wang Y Y, Li W M, Peng Y F, Yuan Q H, Pei X W.De novo transcriptome assembly of common wild rice (Oryza rufipogon Griff.) and discovery of drought-response genes in root tissue based on transcriptomic data. Plos One, 2015, 10: e01314557. |
| [28] | Wang X T, Suo Y Y, Feng Y, Shohag M J I, Gao J, Zhang Q C, Xie S, Lin X Y. Recovery of N-15-labeled urea and soil nitrogen dynamics as affected by irrigation management and nitrogen application rate in a double rice cropping system.Plant Soil, 2011, 343(1-2): 195-208. |
| [29] | Ray S, Dansana P K, Giri J, Deveshwar P, Arora R, Agarwal P, Khurana J P, Kapoor S, Tyagi A K.Modulation of transcription factor and metabolic pathway genes in response to water-deficit stress in rice.Funct Integ Genom, 2011, 11(1): 157-178. |
| [30] | Cheah B H, Nadarajah K, Divate M D, Wickneswari R.Identification of four functionally important microRNA families with contrasting differential expression profiles between drought-tolerant and susceptible rice leaf at vegetative stage.BMC Genom, 2015, 16: 692. |
| [31] | Raorane M L, Pabuayon I M, Varadarajan A R, Mutte S K, Kumar A, Treumann A, Kohli A.Proteomic insights into the role of the large-effect QTL qDTY12.1 for rice yield under drought.Mol Breeding, 2015, 35: 139. |
| [32] | Danquah A, de Zelicourt A, Colcombet J, Hirt H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses.Biotechnol Adv, 2014, 32(1SI): 40-52. |
| [33] | Pouresmael M, Mozafari J.Khavari-nejad R A, Najafi F, Moradi F. Identification of possible mechanisms of chickpea (Cicer arietinum L.) drought tolerance using cDNA-AFLP.J Agric Sci Technol-Iran, 2015, 17(5): 1303-1317. |
| [34] | Paul S, Gayen D, Datta S K, Datta K.Dissecting root proteome of transgenic rice cultivars unravels metabolic alterations and accumulation of novel stress responsive proteins under drought stress.Plant Sci, 2015, 234: 133-143. |
| [35] | Baxter A, Mittler R, Suzuki N.ROS as key players in plant stress signalling.J Exp Bot, 2014, 65(5SI): 1229-1240. |
| [36] | Wang P, Song C.Guard-cell signalling for hydrogen peroxide and abscisic acid.New Phytol, 2008, 178(4): 703-718. |
| [37] | Mittler R, Blumwald E.The roles of ROS and ABA in systemic acquired acclimation.Plant Cell, 2015, 27(1): 64-70. |
| [38] | Mittle R, Vanderauwera S, Suzuki N, Miller G, Tognetti V B, Vandepoele K, Gollery M, Shulaev V, Breusegem F V.ROS signaling: the new wave?Trends Plant Sci, 2011, 16(6): 300-309. |
| [39] | Suzuki N, Miller G, Morales J, Shulaev V, Torres M A, Mittler R.Respiratory burst oxidases: the engines of ROS signaling.Curr Opin Plant Biol, 2011, 14(6): 691-699. |
| [40] | Foyer C H, Noctor G.Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications.Antioxid Redox Sign, 2009, 11(4): 861-905. |
| [41] | Noctor G, Veljovic-Jovanovic S, Driscoll S, Novitskaya L, Foyer C H.Drought and oxidative load in the leaves of C3 plants: a predominant role for photorespiration?Ann Bot, 2002, 89(SI): 841-850. |
| [42] | Kangasjarvi S, Neukermans J, Li S, Aro E, Noctor G.Photosynthesis, photorespiration, and light signalling in defence responses.J Exp Bot, 2012, 63(4SI): 1619-1636. |
| [43] | Zhu Z, Liang Z, Han R.Saikosaponin accumulation and antioxidative protection in drought-stressedBupleurum chinense DC. plants. Environ Exp Bot, 2009, 66(2): 326-333. |
| [44] | Du Y L, Wang Z Y, Fan J W, Turner N C, Wang T, Li F M.β-Aminobutyric acid increases abscisic acid accumulation and desiccation tolerance and decreases water use but fails to improve grain yield in two spring wheat cultivars under soil drying. J Exp Bot, 2012, 63(13): 4849-4860. |
| [45] | Mendelssohn I A, Kleiss B A, Wakeley J S.Factors controlling the formation of oxidized root channels: A review.Wetlands, 1995, 15(1): 37-46. |
| [46] | 苏玲, 章永松, 林咸永. 干湿交替过程中水稻土铁形态和磷吸附解吸的变化. 植物营养与肥料学报, 2001, 7(4) : 410-415. |
| Su L, Zhang Y S, Lin X Y.Changes of iron oxides and phosphorus adsorption-desorption in paddy soils under alternating flooded and dried conditions.Plant Nutr Ferti Sci, 2001, 7(4): 410-415. (in Chinese with English abstract) | |
| [47] | Weber K A, Achenbach L A, Coates J D.Microorganisms pumping iron: anaerobic microbial iron oxidation and reduction.Nat Rev Microbiol, 2006, 4(10): 752-764. |
| [1] | 郭展, 张运波. 水稻对干旱胁迫的生理生化响应及分子调控研究进展[J]. 中国水稻科学, 2024, 38(4): 335-349. |
| [2] | 韦还和, 马唯一, 左博源, 汪璐璐, 朱旺, 耿孝宇, 张翔, 孟天瑶, 陈英龙, 高平磊, 许轲, 霍中洋, 戴其根. 盐、干旱及其复合胁迫对水稻产量和品质形成影响的研究进展[J]. 中国水稻科学, 2024, 38(4): 350-363. |
| [3] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [4] | 伏荣桃, 陈诚, 王剑, 赵黎宇, 陈雪娟, 卢代华. 转录组和代谢组联合分析揭示稻曲病菌的致病因子[J]. 中国水稻科学, 2024, 38(4): 375-385. |
| [5] | 候小琴, 王莹, 余贝, 符卫蒙, 奉保华, 沈煜潮, 谢杭军, 王焕然, 许用强, 武志海, 王建军, 陶龙兴, 符冠富. 黄腐酸钾提高水稻秧苗耐盐性的作用途径分析[J]. 中国水稻科学, 2024, 38(4): 409-421. |
| [6] | 胡继杰, 胡志华, 张均华, 曹小闯, 金千瑜, 章志远, 朱练峰. 根际饱和溶解氧对水稻分蘖期光合及生长特性的影响[J]. 中国水稻科学, 2024, 38(4): 437-446. |
| [7] | 刘福祥, 甄浩洋, 彭焕, 郑刘春, 彭德良, 文艳华. 广东省水稻孢囊线虫病调查与鉴定[J]. 中国水稻科学, 2024, 38(4): 456-461. |
| [8] | 陈浩田, 秦缘, 钟笑涵, 林晨语, 秦竞航, 杨建昌, 张伟杨. 水稻根系和土壤性状与稻田甲烷排放关系的研究进展[J]. 中国水稻科学, 2024, 38(3): 233-245. |
| [9] | 缪军, 冉金晖, 徐梦彬, 卜柳冰, 王平, 梁国华, 周勇. 过量表达异三聚体G蛋白γ亚基基因RGG2提高水稻抗旱性[J]. 中国水稻科学, 2024, 38(3): 246-255. |
| [10] | 尹潇潇, 张芷菡, 颜绣莲, 廖蓉, 杨思葭, 郭岱铭, 樊晶, 赵志学, 王文明. 多个稻曲病菌效应因子的信号肽验证和表达分析[J]. 中国水稻科学, 2024, 38(3): 256-265. |
| [11] | 朱裕敬, 桂金鑫, 龚成云, 罗新阳, 石居斌, 张海清, 贺记外. 全基因组关联分析定位水稻分蘖角度QTL[J]. 中国水稻科学, 2024, 38(3): 266-276. |
| [12] | 魏倩倩, 汪玉磊, 孔海民, 徐青山, 颜玉莲, 潘林, 迟春欣, 孔亚丽, 田文昊, 朱练峰, 曹小闯, 张均华, 朱春权. 信号分子硫化氢参与硫肥缓解铝对水稻生长抑制作用的机制[J]. 中国水稻科学, 2024, 38(3): 290-302. |
| [13] | 周甜, 吴少华, 康建宏, 吴宏亮, 杨生龙, 王星强, 李昱, 黄玉峰. 不同种植模式对水稻籽粒淀粉含量及淀粉关键酶活性的影响[J]. 中国水稻科学, 2024, 38(3): 303-315. |
| [14] | 关雅琪, 鄂志国, 王磊, 申红芳. 影响中国水稻生产环节外包发展因素的实证研究:基于群体效应视角[J]. 中国水稻科学, 2024, 38(3): 324-334. |
| [15] | 许用强, 姜宁, 奉保华, 肖晶晶, 陶龙兴, 符冠富. 水稻开花期高温热害响应机理及其调控技术研究进展[J]. 中国水稻科学, 2024, 38(2): 111-126. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||