
中国水稻科学 ›› 2016, Vol. 30 ›› Issue (5): 469-478.DOI: 10.16819/j.1001-7216.2016.6009
王芳权1,2, 范方军1,2, 李文奇1,2, 朱金燕1,2, 王军1,2, 仲维功1,2, 杨杰1,2,*( )
)
收稿日期:2016-01-18
修回日期:2016-03-03
出版日期:2016-09-10
发布日期:2016-09-10
通讯作者:
杨杰
基金资助:
Fang-quan WANG1,2, Fang-jun FAN1,2, Wen-qi LI1,2, Jin-yan ZHU1,2, Jun WANG1,2, Wei-gong ZHONG1,2, Jie YANG1,2,*( )
)
Received:2016-01-18
Revised:2016-03-03
Online:2016-09-10
Published:2016-09-10
Contact:
Jie YANG
摘要:
利用CRISPR/Cas9技术,针对Pi21基因的两个靶位点(靶位1和靶位2),构建基因敲除载体,转化水稻品种南粳9108。经PCR鉴定,获得了28株T0代阳性转基因植株。对靶位点酶切检测发现,靶位 1突变效率为78.57%,靶位 2突变效率为92.86%;靶位 1和靶位 2同时突变的效率为78.57%,突变效率较高。通过对靶位点进行测序,发现靶位点突变类型较多,包括碱基缺失、碱基插入、碱基缺失后插入其他碱基和大片段DNA缺失等类型。对突变株系进行氨基酸预测,发现大部分株系都存在移码突变现象而使基因突变彻底,少数株系表现为部分氨基酸的缺失或变异。成功敲除了水稻Pi21基因,并对突变效率和类型进行了分析,为进一步验证Pi21基因功能、培育广谱抗稻瘟病的水稻新品系奠定了基础。
中图分类号:
王芳权, 范方军, 李文奇, 朱金燕, 王军, 仲维功, 杨杰. 利用CRISPR/Cas9技术敲除水稻Pi21基因的效率分析[J]. 中国水稻科学, 2016, 30(5): 469-478.
Fang-quan WANG, Fang-jun FAN, Wen-qi LI, Jin-yan ZHU, Jun WANG, Wei-gong ZHONG, Jie YANG. Knock-out Efficiency Analysis of Pi21 Gene Using CRISPR/Cas9 in Rice[J]. Chinese Journal OF Rice Science, 2016, 30(5): 469-478.
| 引物名称 Primer name | 引物序列(5'→3') Primer sequence (5'→3') |
|---|---|
| Pi21g-F | ACCAAAGCCTGTCTATCTGC |
| Pi21g-R | CACATCGATCAGCCTCGTG |
| Pi21UT1-F | GGCAGAAGCTGTGCAAGAAGATC |
| Pi21UT1-R | AAACGATCTTCTTGCACAGCTTC |
| Pi21UT2-F | GCCGCCGCTGCGATGCCAAGATC |
| Pi21UT2-R | AAACGATCTTGGCATCGCAGCGG |
| pU-F | CTCCGTTTTACCTGTGGAATCG |
| gRNA-R | CGGAGGAAAATTCCATCCAC |
| Pi21T1L-F | TTCAGAGGTCTCTCTCGCACTGGAATCGGCAGCAAAGG |
| Pi21T1L-R | AGCGTGGGTCTCGTCAGGGTCCATCCACTCCAAGCTC |
| Pi21T2L-F | TTCAGAGGTCTCTCTGACACTGGAATCGGCAGCAAAGG |
| Pi21T2L-R | AGCGTGGGTCTCGACCGGGTCCATCCACTCCAAGCTC |
| SP1 | CCCGACATAGATGCAATAACTTC |
| SP2 | GCGCGGTGTCATCTATGTTA |
| Pi21T1-F | AGGCTAATCAGCAGTGTTCCT |
| Pi21T1-R | CAGCTTGCACTCCGGCTTCG |
| Pi21T2-F | ATTGGTAACATTCGGCAAATT |
| Pi21T2-R | GTTCTTCACGTCGTACTCCA |
表1 本研究所用的引物
Table 1 Primers used in this research.
| 引物名称 Primer name | 引物序列(5'→3') Primer sequence (5'→3') |
|---|---|
| Pi21g-F | ACCAAAGCCTGTCTATCTGC |
| Pi21g-R | CACATCGATCAGCCTCGTG |
| Pi21UT1-F | GGCAGAAGCTGTGCAAGAAGATC |
| Pi21UT1-R | AAACGATCTTCTTGCACAGCTTC |
| Pi21UT2-F | GCCGCCGCTGCGATGCCAAGATC |
| Pi21UT2-R | AAACGATCTTGGCATCGCAGCGG |
| pU-F | CTCCGTTTTACCTGTGGAATCG |
| gRNA-R | CGGAGGAAAATTCCATCCAC |
| Pi21T1L-F | TTCAGAGGTCTCTCTCGCACTGGAATCGGCAGCAAAGG |
| Pi21T1L-R | AGCGTGGGTCTCGTCAGGGTCCATCCACTCCAAGCTC |
| Pi21T2L-F | TTCAGAGGTCTCTCTGACACTGGAATCGGCAGCAAAGG |
| Pi21T2L-R | AGCGTGGGTCTCGACCGGGTCCATCCACTCCAAGCTC |
| SP1 | CCCGACATAGATGCAATAACTTC |
| SP2 | GCGCGGTGTCATCTATGTTA |
| Pi21T1-F | AGGCTAATCAGCAGTGTTCCT |
| Pi21T1-R | CAGCTTGCACTCCGGCTTCG |
| Pi21T2-F | ATTGGTAACATTCGGCAAATT |
| Pi21T2-R | GTTCTTCACGTCGTACTCCA |

图1 Pi21基因编码序列比对 南粳9108感稻瘟病, 为Pi21感病基因型; Owarihatamochi抗稻瘟病, 为pi21抗病基因型。
Fig. 1. Coding sequences co-linearity analysis of Pi21 gene. Nanjing 9108 is a blast sensitive variety, containing the susceptible allele Pi21. Owarihatamochi is a blast resistant variety, containing the resistant allele pi21.
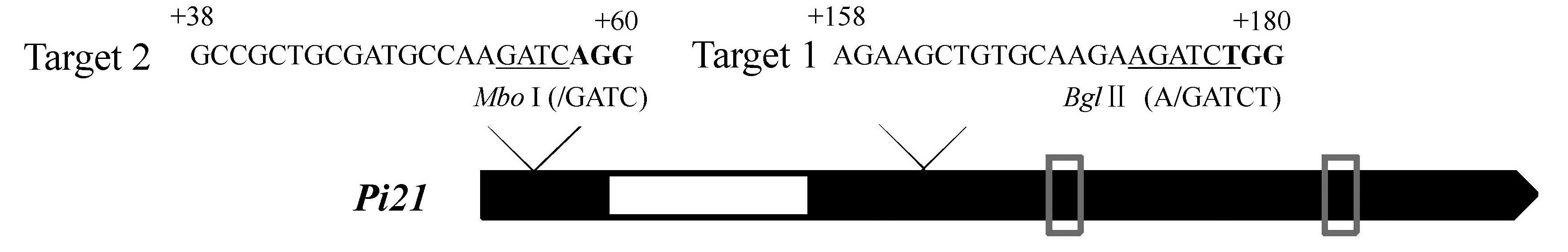
图2 Pi21基因结构及gRNA靶点位置 加粗的碱基为PAM,下划线标出的碱基为限制性内切酶识别位点,黑色区域为外显子,白色区域为内含子,灰色方框标识区域为富脯氨酸元件。
Fig. 2. Schematic diagram of Pi21 gene structure and gRNA targets. The base in bold indicates PAM; The restriction endonuclease recognized site is underlined; The black region indicates exon; The white region indicates intron; The gray box region indicates proline-rich motifs.
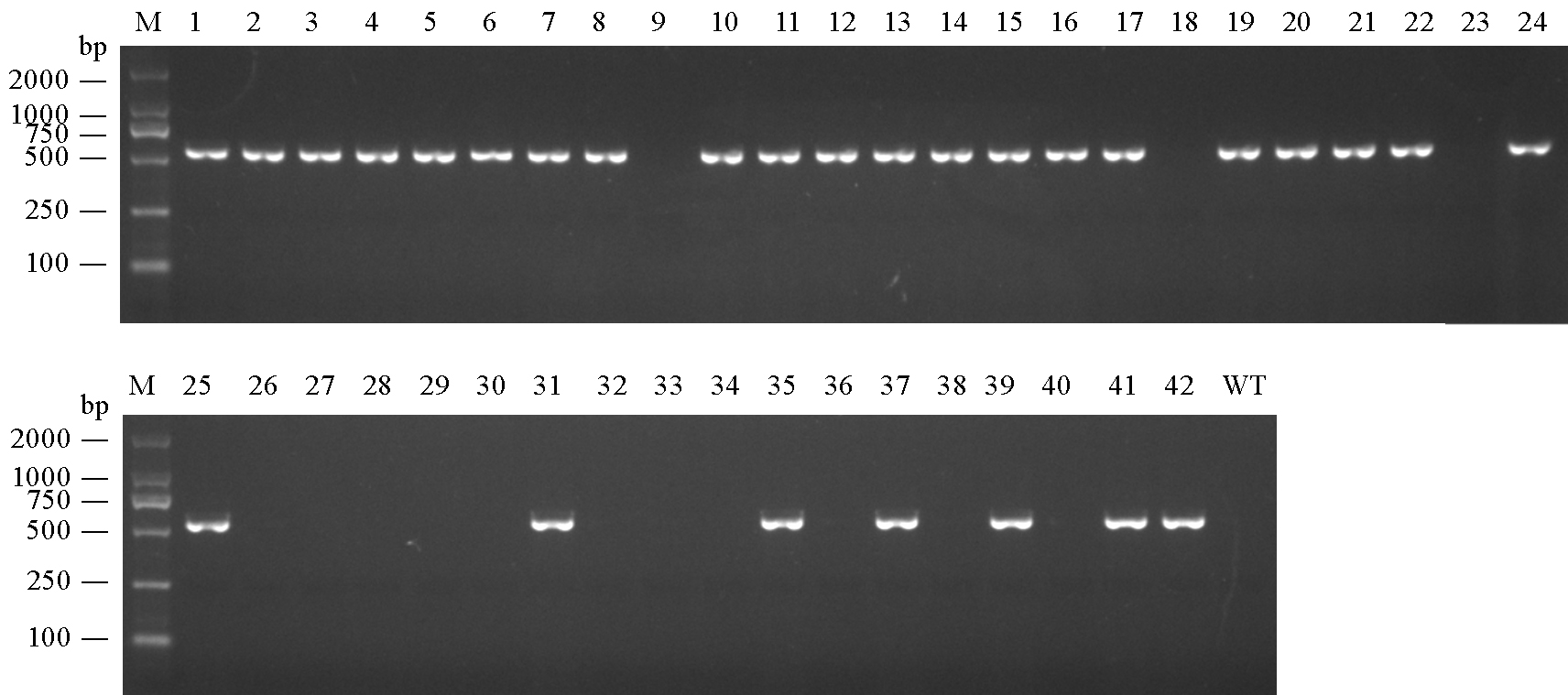
图4 PCR鉴定转基因阳性植株 M-DL 2000 DNA标记; 1~42-T0代转基因株系; WT-南粳9108(野生型)。
Fig. 4. PCR identification of the positive transgenic plants. M, DL 2000 DNA marker; 1-42, T0 transgenic lines; WT, Nanjing 9108 (wild-type).
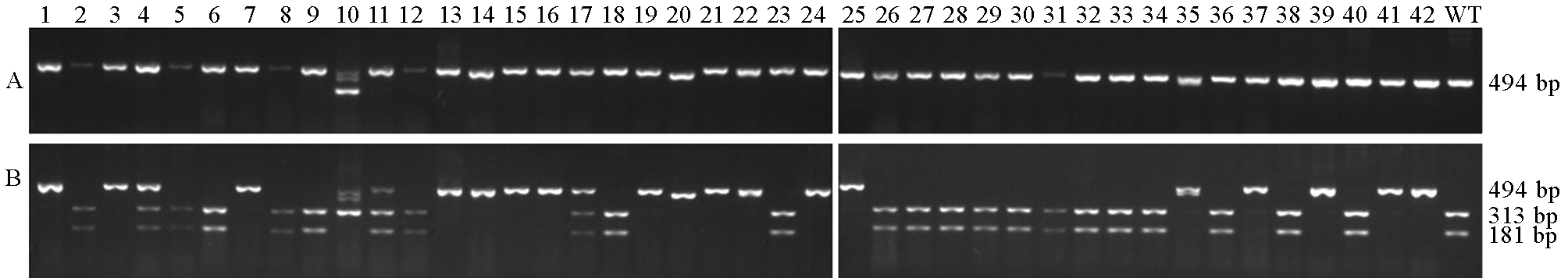
图5 酶切鉴定靶位 1突变情况 A-酶切前; B-酶切后。 1~42-T0代转基因株系; WT-南粳9108(野生型)。
Fig. 5. Mutation identification at Target 1 by enzyme digestion. A, Before digestion; B, After digestion. 1~42, T0 transgenic lines; WT, Nanjing 9108 (wild-type).
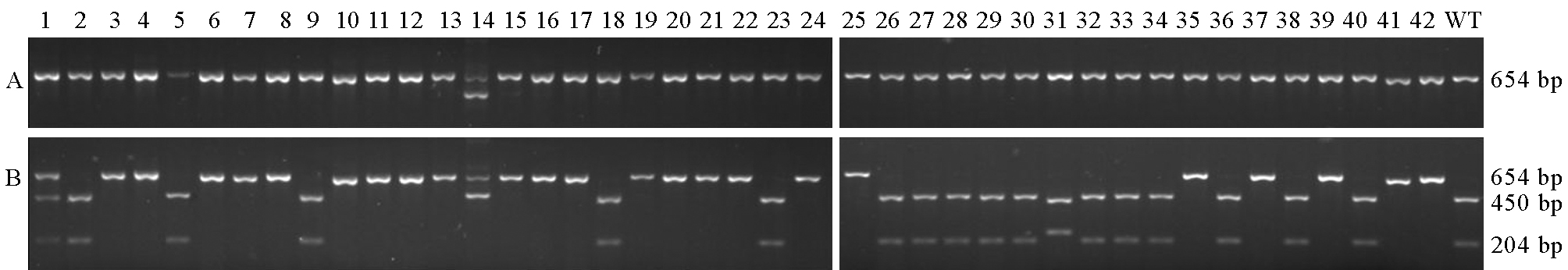
图6 酶切鉴定靶位 2突变情况 A-酶切前; B-酶切后。 1~42-T0代转基因株系; WT-南粳9108(野生型)。
Fig. 6. Mutation identification at Target 2 by enzyme digestion. A, Before digestion; B, After digestion. 1~42, T0 transgenic lines; WT, Nanjing 9108 (wild-type).

图8 靶位点突变序列分析 “-1”和“-2”表示不同突变类型,括号内的数值为测序的克隆数,箭头所指的位置为Cas9剪切位点,加粗碱基为前间区序列邻近基序(PAM),红色字体的碱基为靶序列,蓝色字体的碱基为插入碱基。WT, 野生型;#, 转基因株系; d, 缺失; i, 插入。
Fig. 8. Analysis of mutation sequences of targets. “-1” and “-2” indicate different mutation type; The numbers in brackets are the numbers of sequenced clone; The arrows indicate the digestion sites of Cas9; The base in bold indicates protospacer adjacent motif(PAM); The base in red is the target sequence; The base in blue is the insert base; WT, Wild-type; #, Transgenic lines; d, Deletion; i, Insert.
| [1] | Belhaj K, Chaparro-Garcia A, Kamoun S, et al.Editing plant genomes with CRISPR/Cas9.Curr Opin Biotech, 2015, 32: 76-84. |
| [2] | Baltes N J, Voytas D F.Enabling plant synthetic biology through genome engineering.Trends Biotechnol, 2015, 33(2): 120-131. |
| [3] | 解莉楠, 宋凤艳, 张旸. CRISPR/Cas9 系统在植物基因组定点编辑中的研究进展. 中国农业科学, 2015, 48(9): 1669-1677. |
| Xie L N, Song F Y, Zhang Y.Progress in research of CRISPR/Cas9 system in genome targeted editing in plants.Sci Agric Sin, 2015, 48(9): 1669-1677. (in Chinese with English abstract) | |
| [4] | 李君, 张毅, 陈坤玲, 等. CRISPR/Cas 系统: RNA 靶向的基因组定向. 遗传, 2013, 35(11): 1265-1273. |
| Li J, Zhang Y, Chen K L, et al.CRISPR/Cas: A novel way of RNA-guided genome editing.Hereditas, 2013, 35(11): 1265-1273. (in Chinese with English abstract) | |
| [5] | Jiang W, Bikard D, Cox D, et al.RNA-guided editing of bacterial genomes using CRISPR-Cas systems.Nat Biotechnol, 2013, 31(3): 233-239. |
| [6] | DiCarlo J E, Norville J E, Mali P, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems.Nucleic Acids Res, 2013, 41(7): 4336-4343. |
| [7] | Cong L, Ran F A, Cox D, et al.Multiplex genome engineering using CRISPR/Cas systems.Science, 2013, 339(6121): 819-823. |
| [8] | Mali P, Yang L, Esvelt K M, et al.RNA-guided human genome engineering via Cas9.Science, 2013, 339(6121): 823-826. |
| [9] | Wang H, Yang H, Shivalila C S, et al.One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering.Cell, 2013, 153(4): 910-918. |
| [10] | Feng Z, Zhang B, Ding W, et al.Efficient genome editing in plants using a CRISPR/Cas system.Cell Res, 2013, 23(10): 1229-1232. |
| [11] | Jiang W, Zhou H, Bi H, et al.Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice.Nucleic Acids Res, 2013, 41(20): e188. |
| [12] | Mao Y, Zhang H, Xu N, et al.Application of the CRISPR-Cas system for efficient genome engineering in plants.Mol Plant, 2013, 6(6): 2008-2011. |
| [13] | Shan Q, Wang Y, Li J, et al.Targeted genome modification of crop plants using a CRISPR-Cas system.Nat Biotechnol, 2013, 31(8): 686-688. |
| [14] | Zhang H, Zhang J, Wei P, et al.The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation.Plant Biotechnol J, 2014, 12(6): 797-807. |
| [15] | Li J F, Norville J E, Aach J, et al.Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9.Nat Biotechnol, 2013, 31(8): 688-691. |
| [16] | Nekrasov V, Staskawicz B, Weigel D, et al.Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease.Nat Biotechnol, 2013, 31(8): 691-693. |
| [17] | Liang Z, Zhang K, Chen K, et al.Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system.J Genet Genom, 2014, 41(2): 63-68. |
| [18] | Wang Y, Cheng X, Shan Q, et al.Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew.Nat Biotechnol, 2014, 32(9): 947-951. |
| [19] | Jacobs T B, LaFayette P R, Schmitz R J, et al. Targeted genome modifications in soybean with CRISPR/Cas9.BMC Biotechnol, 2015, 15(1): 16. |
| [20] | Li Z, Liu Z B, Xing A, et al.Cas9-guide RNA directed genome editing in soybean.Plant Physiol, 2015, 169(2): 960-970. |
| [21] | Sun X, Hu Z, Chen R, et al.Targeted mutagenesis in soybean using the CRISPR-Cas9 system.Sci Rep-UK, 2015, 5: 10342. |
| [22] | Brooks C, Nekrasov V, Lippman Z B, et al.Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system.Plant Physiol, 2014, 166(3): 1292-1297. |
| [23] | Miao J, Guo D, Zhang J, et al.Targeted mutagenesis in rice using CRISPR-Cas system.Cell Res, 2013, 23(10): 1233-1236. |
| [24] | Shan Q, Wang Y, Li J, et al.Genome editing in rice and wheat using the CRISPR/Cas system.Nat Protec, 2014, 9(10): 2395-2410. |
| [25] | Xu R F, Li H, Qin R Y, et al.Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system.Sci Rep-UK, 2015, 5: 11491. |
| [26] | Ma X, Zhang Q, Zhu Q, et al.A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants.Mol Plant, 2015, 8(8): 1274-1284. |
| [27] | 鄂志国, 王磊. 水稻抗病性基因的克隆和功能研究进展. 遗传, 2009, 31(10): 999-1005. |
| E Z G, Wang L. Advance on the cloning and functional analysis of disease resistance genes in rice.Hereditas, 2009, 31(10): 999-1005. (in Chinese with English abstract) | |
| [28] | Fukuoka S, Okuno K.QTL analysis and mapping of pi21, a recessive gene for field resistance to rice blast in Japanese upland rice.Theor Appl Genet, 2001, 103(2): 185-190. |
| [29] | Fukuoka S, Saka N, Koga H, et al.Loss of function of a proline-containing protein confers durable disease resistance in rice.Science, 2009, 325(5943): 998-1001. |
| [30] | 朱金燕, 王军, 范方军, 等. 水稻稻瘟病广谱抗病新等位基因pi21t的鉴定及其抗性应用. 华北农学报, 2014, 29(6): 11-15. |
| Zhu J Y, Wang J, Fan F J.et al.Identification and application of one new rice blast broad-spectrum resistance allele pi21t.Acta Agric Bor-Sin, 2014, 29(6): 11-15. (in Chinese with English abstract) | |
| [31] | 王才林, 张亚东, 朱镇, 等. 优良食味粳稻新品种南粳9108的选育与利用. 江苏农业科学, 2013, 41(9): 86-88. |
| Wang C L, Zhang Y D, Zhu Z, et al.Breeding and application of Nanjing 9108, a new japonica variety with excellent eating quality.Jiangsu Agric Sci, 2013, 41(9): 86-88. | |
| [32] | Xie K, Yang Y.RNA-guided genome editing in plants using a CRISPR-Cas system.Mol Plant, 2013, 6(6): 1975-1983. |
| [33] | 朱立宏. 关于我国水稻高产育种的我见. 南京农业大学学报, 2007, 30(1): 129-135. |
| Zhu L H.Some critical considerations on rice high-yielding breeding in China.J Nanjing Agric Univ, 2007, 30(1): 129-135. (in Chinese with English abstract) | |
| [34] | 凌忠专, 雷财林, 王久林. 稻瘟病菌生理小种研究的回顾与展望. 中国农业科学, 2004, 37(12): 1849-1859. |
| Ling Z Z, Lei C L, Wang J L.Review and prospect for study of physiologic races on rice blast fungus (Pyricularia grasea).Sci Agric Sin, 2004, 37(12): 1849-1859. (in Chinese with English abstract) |
| [1] | 许丹洁, 林巧霞, 李正康, 庄小倩, 凌宇, 赖美玲, 陈晓婷, 鲁国东. OsOPR10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2024, 38(4): 364-374. |
| [2] | 童琪, 王春燕, 阙亚伟, 肖宇, 王政逸. 稻瘟病菌热激蛋白(HSP)40编码基因MoMHF6的鉴定及功能研究[J]. 中国水稻科学, 2023, 37(6): 563-576. |
| [3] | 陈明亮, 熊文涛, 沈雨民, 熊焕金, 罗世友, 吴小燕, 胡兰香, 肖叶青. 广谱抗稻瘟病水稻保持系赣香B的抗性遗传解析[J]. 中国水稻科学, 2023, 37(5): 470-477. |
| [4] | 李景芳, 温舒越, 赵利君, 陈庭木, 周振玲, 孙志广, 刘艳, 陈海元, 张云辉, 迟铭, 邢运高, 徐波, 徐大勇, 王宝祥. 基于CRISPR/Cas9技术创制耐盐香稻[J]. 中国水稻科学, 2023, 37(5): 478-485. |
| [5] | 李刚, 高清松, 李伟, 张雯霞, 王健, 程保山, 王迪, 高浩, 徐卫军, 陈红旗, 纪剑辉. 定向敲除SD1基因提高水稻的抗倒性和稻瘟病抗性[J]. 中国水稻科学, 2023, 37(4): 359-367. |
| [6] | 段敏, 谢留杰, 高秀莹, 唐海娟, 黄善军, 潘晓飚. 利用CRISPR/Cas9技术创制广亲和水稻温敏雄性不育系[J]. 中国水稻科学, 2023, 37(3): 233-243. |
| [7] | 王雨, 孙全翌, 杜海波, 许志文, 吴科霆, 尹力, 冯志明, 胡珂鸣, 陈宗祥, 左示敏. 利用抗稻瘟病基因Pigm和抗纹枯病数量性状基因qSB-9TQ、qSB-11HJX改良南粳9108的抗性[J]. 中国水稻科学, 2023, 37(2): 125-132. |
| [8] | 王石光, 陆展华, 刘维, 卢东柏, 王晓飞, 方志强, 巫浩翔, 何秀英. 应用CRISPR/Cas9技术与分子标记辅助选择创制广东丝苗米新种质[J]. 中国水稻科学, 2023, 37(1): 29-36. |
| [9] | 张元野, 尹丽颖, 李荣田, 何明良, 刘欣欣, 潘婷婷, 田晓杰, 卜庆云, 李秀峰. 利用CRISPR/Cas9技术创制Rc基因恢复红稻[J]. 中国水稻科学, 2022, 36(6): 572-578. |
| [10] | 尹丽颖, 张元野, 李荣田, 何明良, 王芳权, 许扬, 刘欣欣, 潘婷婷, 田晓杰, 卜庆云, 李秀峰. 利用CRISPR/Cas9技术创制高效抗除草剂水稻[J]. 中国水稻科学, 2022, 36(5): 459-466. |
| [11] | 周永林, 申小磊, 周立帅, 林巧霞, 王朝露, 陈静, 冯慧捷, 张振文, 陈晓婷, 鲁国东. OsLOX10正调控水稻对稻瘟病和白叶枯病的抗性[J]. 中国水稻科学, 2022, 36(4): 348-356. |
| [12] | 李兆伟, 孙聪颖, 零东兰, 曾慧玲, 张晓妹, 范凯, 林文雄. 利用CRISPR/Cas9创建osarf7突变体及其农艺性状调查[J]. 中国水稻科学, 2022, 36(3): 237-247. |
| [13] | 梁敏敏, 张华丽, 陈俊宇, 戴冬青, 杜成兴, 王惠梅, 马良勇. 利用CRISPR/Cas9技术创制抗稻瘟病香型早籼温敏核不育系[J]. 中国水稻科学, 2022, 36(3): 248-258. |
| [14] | 曹煜东, 肖湘谊, 叶乃忠, 丁晓雯, 易晓璇, 刘金灵, 肖应辉. 生长素调控因子OsGRF4协同调控水稻粒形和稻瘟病抗性[J]. 中国水稻科学, 2021, 35(6): 629-638. |
| [15] | 刘树芳, 董丽英, 李迅东, 周伍民, 杨勤忠. 持有Pi9基因的水稻单基因系IRBL9-W对稻瘟病菌苗期和成株期抗性差异[J]. 中国水稻科学, 2021, 35(3): 303-310. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||